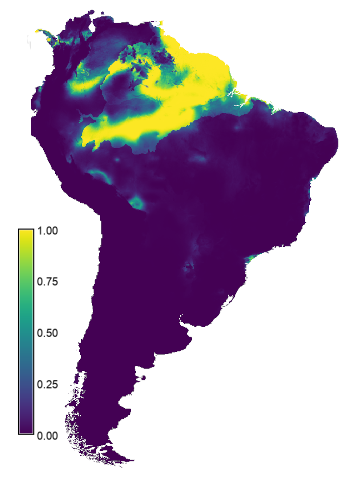
NAME
r.maxent.predict - Use a Maxent model to create a suitability distribution layerKEYWORDS
modeling, MaxentSYNOPSIS
Flags:
- -e
- Automatically adapt resolution
- When the ns and ew resolution are not the same, nearest neighbor resampling will be used to ensure both are the same.
- -c
- Do not apply clamping
- Do not apply clamping when projecting.
- -f
- Fade effect clamping
- Reduce prediction at each point in projections by the difference between clamped and non-clamped output at that point.
- -i
- Copy maxent.jar to addon directory
- Copy the maxent.jar (path provided with the 'maxent' parameter) to the addon scripts directory.
- -u
- Overwrites maxent.jar in addon directory
- Copy the maxent.jar (path provided with the 'maxent' parameter) to the addon scripts directory. If the file already exists in the addon directory, it is overwritten.
- -p
- Print Maxent command
- Print the Maxent command used to create the prediction layer. For debugging.
- --overwrite
- Allow output files to overwrite existing files
- --help
- Print usage summary
- --verbose
- Verbose module output
- --quiet
- Quiet module output
- --ui
- Force launching GUI dialog
Parameters:
- lambda=name [required]
- Lambda model file
- Lambda model file created by Maxent or the r.maxent.train addon.
- raster=name[,name,...]
- Names of the input raster layers
- Names of the raster layers representing the environmental variables used in the Maxent model.
- variables=string
- variable names
- Names of the environmental parameter(s) as used in the model. These need to be given in the same order as the corresponding raster layers. If left out, the names are assumed to be the same as the name of the raster layers.
- alias_file=name
- csv file with variable and layer names
- A csv file with in the first column the names of the explanatory variables used in the model, and in the second column the names of corresponding raster layers. Make both are provided in the same order.
- output=name [required]
- Prediction raster layer
- The name of the raster layer with the predicted suitability scores
- maxent=name
- Location Maxent jar file
- Give the path to the Maxent executable file (maxent.jar)
- memory=memory in MB
- Maximum memory to be used (in MB)
- Maximum memory to be used by Maxent (in MB)
- Default: 300
Table of contents
DESCRIPTION
The r.maxent.predict addon can be used to apply a previously calculated Maxent model to a new set of environmental raster data. It requires a .lambdas file describing a Maxent model, and the name of raster layers for all predictor variables described in the lambdas file. The .lambdas file can be created by the r.maxent.train addon, or by the Maxent software directly.For convenience, r.maxent.train creates a file maxent_explanatory_variable_names.csv, which you can check for the names of the predictor variables. If these are different from the input raster layers, you can provide the variable names using the variables parameter. Alternatively, you can provide a CSV file with the names of the explanatory variables (first column) and the names of the corresponding raster layers (second column).
Maxent includes the option of “clamping” projections. This constrains the values for environmental values in the projected range to the limit of that variable that is found in the training range. By default, Maxent applied clamping. You can disable this by setting the -c flag. You also have the option to reduce the prediction at each point in projections by the difference between clamped and non-clamped output at that point. Use the -f to enable this option.
NOTES
This addon requires the Maxent software. You can download the software from the Maxent website. The software includes a Maxent.jar file. The addon expects a copy of the executable in the GRASS GIS addon directory. If it is not there, you need to provide the path to the maxent.jar file using the maxent parameter. To avoid having to provide this each time again, you can use the -i flag. If set, the maxent.jar file will be copied to the addon/script directory. Note that if you already did this when running r.maxent.train, there is no need to repeat it here.If you want to update the Maxent.jar file, use the -u flag. Removing the Maxent.jar file needs to be done manually. Go to the GRASS GIS addon directory, and delete the Maxent.jar file. To find the addon directory, open GRASS GIS, and type `echo $GRASS_ADDON_BASE` on the command line.
Examples
The examples below use a dataset that you can download from here. It includes a vector point layer with observation locations of the pale-throated sloth (Bradypus tridactylus) from GBIF, the IUCN RedList range map of the species, a boundary layer of the South American countries from NaturalEarth and a number of bioclim raster layers from WorldClim version 2.1, representing the climate conditions representing the period 1970-2000 and the climate conditions predicted for 2061–2080 based on the GCM BCC-CSM2-MR and SSP 585.
The zip file contains a folder sampledata. This is a location with five subfolders PERMANENT, sloth, current, future and model01. Copy this Location to a GRASS Database (use an existing one or create one first). If you are not familiar with the concept of Locations and Mapsets, please first read the explanation about the GRASS GIS database.
Unzip the file, start up GRASS GIS, open the GRASS GIS database to which you copied the folder sampledata, switch to the Location sampledata and open the mapset model01. This mapset should have access to the other mapsets. This addon is part of a series of three addons that can be used to prepare the data, train a maxent presence only model, and to use the model to create prediction layers.

A workflow, from data preparation, training a model to model prediction using three GRASS GIS addons.
The examples below show how to use the three addons in sequence. Only the basic options are shown. For a detailed account of all options, check out the Maxent tutorial on the Maxent website.
1: Data preparation
You can use the v.maxent.swd to create the required input layers. The code below creates the SWD file with the locations where the species has been recorded (species_output) and a SWD file with randomly created background point locations (bgr_ouput). The SWD files contain, for each location, the values of the raster layers selected with the evp_maps parameter. With the parameter export_rasters you tell the addon to export the raster layers as well.v.maxent.swd -t \ species=Bradypus_tridactylus \ evp_maps=bio02,bio03,bio08,bio09,bio13,bio15,bio17 \ evp_cat=sa_eco_l2@current \ alias_cat=landuse \ nbgp=10000 \ bgr_output=bgrd_swd.csv \ species_output=spec_swd.csv \ export_rasters=envlayers
The output is a folder with the so-called SWD files with the XY coordinates for the species presence location (spec_swd.csv) and the background locations (bgrd_swd.csv. Both also include the values of the input raster layers for the given point locations. In addition, there is the subfolder envlayers with the environmental raster layers in ascii format.
2: Train the model
Use the output of v.maxent.swd as input for r.maxent.train. First create a subfolder output_model1, so we can write the output to that folder.The projectionlayers parameter is optionally. If you set it, a raster prediction layer will be created that represent the potential suitability distribution under current conditions (the conditions used to train the model).
With the -y and -b flags the point layers with the sample predictions and the predictions at the background point locations are created. Their values correspond to the values of the raster prediction layer.
r.maxent.train -y -b -g \ samplesfile=spec_swd.csv \ environmentallayersfile=bgrd_swd.csv \ togglelayertype=landuse \ projectionlayers=envlayers \ samplepredictions=model_1_samplepred \ backgroundpredictions=model_1_bgrdpred \ predictionlayer=model_1_suitability_current \ outputdirectory=output_model1
When r.maxent.train is finished, go to the output folder and open the Bradypus_tridactylus.html file for an explanation of the different model outputs and model evaluation statistics. For a more detailed explanation, see the tutorial on the Maxent website.
In your current mapset, you'll find the raster prediction layer, and the sample and background point layers with the predicted values.
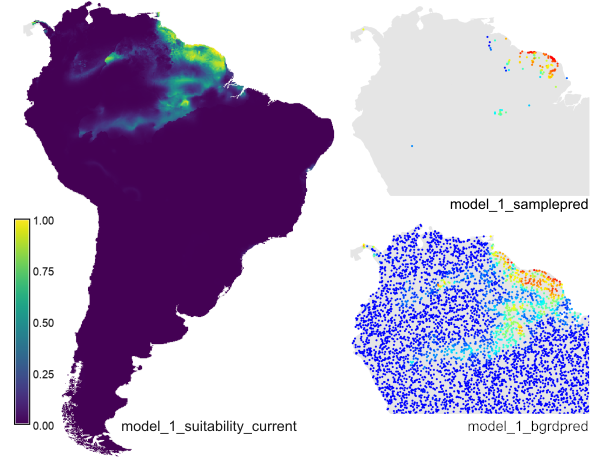
The example creates the prediction raster layer 'model_1_suitability_current', the sample point layer 'model_1_samplepred' and the background point layer 'model_bgrdpred' (for the latter, only part of the map is shown here).
3: Create a prediction layer
The third step is to use the model created in the previous step to predict the species suitability distribution under future climates. Note, we are going to make the (unrealistic) assumption that the ecosystems do not change.r.maxent.predict lambda=output_model1/Bradypus_tridactylus.lambdas raster=bio02_ssp585,bio03_ssp585,bio08_ssp585,bio09_ssp585,bio13_ssp585,bio15_ssp585,bio17_ssp585,sa_eco_l2@current variables=bio02,bio03,bio08,bio09,bio13,bio15,bio17,landuse \ output=model_1_ssp585
The resulting layer is written to the current mapset as model_1_ssp585 (right map in the figure below). The results suggest the area with suitable conditions will increase under future climates compared the that under the current conditions (left map in the figure below). Not sure that is realistic.
REFERENCES
- Steven J. Phillips, Miroslav Dudík, Robert E. Schapire. 2020: Maxent software for modeling species niches and distributions (Version 3.4.1). Available from url: https://biodiversityinformatics.amnh.org/open_source/maxent and https://github.com/mrmaxent/Maxent
- Steven J. Phillips, Miroslav Dudík, Robert E. Schapire. 2004: A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, pages 655-662, 2004.
- Steven J. Phillips, Robert P. Anderson, Robert E. Schapire. 2006: Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190:231-259, 2006.
- Jane Elith, Steven J. Phillips, Trevor Hastie, Miroslav Dudík, Yung En Chee, Colin J. Yates. 2011: A statistical explanation of MaxEnt for ecologists. Diversity and Distributions, 17:43-57, 2011.
SEE ALSO
- v.maxent.swd, creating species and background swd files and prediction rasters that can be used directly by the r.maxent.train addon (or the Maxent software itself) to create species distribution models.
- r.out.maxent_swd, creating species and background swd files based on species distribution data in raster format.
- r.maxent.train, creates a maxent model based on presence point data a set of environmental predictor layers.
AUTHOR
Paulo van Breugel, https://ecodiv.earth
HAS green academy University of Applied Sciences
Innovative
Biomonitoring research group
Climate-robust
Landscapes research group
SOURCE CODE
Available at: r.maxent.predict source code (history)
Latest change: Friday Feb 23 08:34:21 2024 in commit: d0e4d42ad622951cadf3bdcde73140d0c6b92199
Main index | Raster index | Topics index | Keywords index | Graphical index | Full index
© 2003-2024 GRASS Development Team, GRASS GIS 8.3.3dev Reference Manual