
Introduction to GRASS GIS Software

by
Markus Neteler
Hannover - Germany
2. Edition, Feb. 1998
(minor HTML-corrections May 1998)
This text is based on "Introduction to GRASS GIS software" written by the author for
Institute of Remote Sensing
Anna University
Chennai (Madras) – India
within the scope of a
Ongoing GTZ project
in March 1997
© Markus Neteler, Hannover 1997-98
Author's address:
Markus Neteler
Institute of Physical Geography and Landscape Ecology
University of Hannover
Germany
email: neteler@itc.it
Preface
The second edition of "Introduction to GRASS GIS Software" was necessary to update the document to the new developments in GRASS. In 1997 the GRASS Research Group was formed at Baylor University, Waco (Texas), U.S.A. which now maintaines the GRASS software package. Because of the need to keep GRASS available for the large user community worldwide, the GRASS Research Group has taken over support, research, and development from USA-CERL with the intent of keeping GRASS public-domain. The following project parts are established: The development of new GRASS versions (floating point GRASS is planned for summer 1998), new documentation, and also web-based information in internet.
This document was slightly changed and updated to new GRASS structures (mainly xy-locations). The authors hopes to give an easy introduction to this nice software package.
Markus Neteler
Hannover, 26. Feb. 1998
Table of Contents
I. INTRODUCTION
*1. General Introduction
*1.1 System requirement
*1.2 General structure of GRASS
*1.3 Major modules in GRASS
*2. Data formats
*2.1 Raster data
*2.2 Vector data
*2.3 Point data
*3. Map output
*II. DATABASE CREATION
*1. The first steps
*1.1 Projections and coordinate systems
*1.2 What information are necessary ?
*2. Definition of a database
*1.1 Locations and mapsets
*2.2 The Projections and coordinate systems
*III. General file management commands
*1. Introduction
*2. Help system and file commands
*3. Deleting of mapsets and locations
*IV. Point Data Management
*1. Introduction to point data management
*1.1 Import and export of sites
*1.2 Display of the sites - displaycommands
*1.3 A few words about the GRASS monitors
*2. DTM analysis
*2.1 Topological analysis
*2.2 Interpolation of surfaces from spot heights
*2.3 Statistical report on sites files
*3.
Triangulation *3.1 Delaunay triangulation
*3.2 Thiessen polygons
*4. Conversions of sites to other formats
*4.1 Sites to raster format
*4.2 Sites to vector format
*V. VECTOR DATA MANAGEMENT
*1. Introduction to vector data management
*1.1 Import of vector data
*1.2 Export of vectordata
*2. Digitizing in GRASS
*2.1 Import of a scanned map
*2.2 Digitizing
*3. Conversion of vector layers to other formats
*3.1 Vector format to sites format
*3.2. Vector format to raster format
*4. Overlay and splitting of vectorlayers
*VI. RASTER DATA MANAGEMENT
*1. Introduction to raster data management
*1.1 Import and export of rasterdata
*2. Colortable modifications
*2.1 Assign a standard colortable
*2.2 Change the colortable interactively
*2.3 Display of colortable
*3. Display of raster maps in different scales
*4. Query raster cell contents and display of profiles
*5. Classification of raster maps
*6. Rescaling of raster maps
*7. Managing label information in raster maps
*8. Display of histograms and cell distributions
*9. Buffering
*10. Example: Creating a potential soil erosion map
*10.1 Generating the slope map
*10.2 Generating the vegetation cover map
*10.3 Generation the erodibility map
*10.4 Classification of the input maps
*10.5. Validation of the result
*11. Overview of the functions in r.mapcalc
*12. Overview of GRASS capabilities in watershed modelling
*12.1 Introduction
*12.2 Watershed calculation and creation
*12.3 DEM Calculations
*12.4 Rainfall data preparation
*12.5 Erosion and transportation models
*12.6 Runoff calculation
*12.7 Calculations on raster layers
*12.8 Automated boundary and line digitizing
*VII. IMAGE PROCESSING
*1. Introduction
*1.1 Definition of the xy-location for unrectified images
*1.2 Import of satellite images
*2. Rectification of satellite images and aerial photos
*2.1 Collect the image bands in a group
*2.2 Specifying the target location
*2.3 Setting the ground control points (GCPs)
*2.4 Verifying the quality of GCPs
*2.5 Minimum number of GCPs
*2.6 Transforming the images
*3. Image enhancements
*3.1 Contrast stretch
*3.2 Principal and canonical component analysis
*3.3 Fourier and inverse Fourier transformation
*3.4 Matrix filters
*4. Color composites
*5. Calculation of band ratios
*6. Introduction to satellite image classification
*6.1 Spectral pattern recognition
*6.2 Spatial pattern recognition
*7. Enhancement of image resolution using IHS/RGB transformation – image fusion
*VIII. CONCLUSION
*APPENDIX
*Appendix 1: Frequently asked questions and their answers
*Appendix 2: Script programming in GRASS
*Appendix 3: GRASS Command Overview
*GRASS display modules
*GRASS file management modules
*GRASS imagery modules
*GRASS data import/processing modules
*GRASS paint/print output modules
*GRASS raster modules
*GRASS sites management modules
*GRASS vector modules
*GRASS shell scripts
*Appendix 4: Linux Hardware compatibility list
*Appendix 5: Internet sites for GRASS
*Appendix 6: GRASS on CDROM
*References
*
The general purpose of using geographical information systems (GIS) is to collect, analyse, manipulate and display spatial data with their associated attributes. During the last decades a variety of GIS have been developed. Private companies as well as governmental, municipal organizations and universities are using these systems. Geographical information systems offer a simple and efficient way of spatial analysis.
A powerful GIS-package is the GRASS-Software. The name of GRASS-GIS is an abbreviation for "Geographic Resource Analysis Support System". It is a hybrid GIS-package - it manages raster, vector as well as point data and contains image processing modules. GRASS was released to public in the year 1989. It is a development of the U.S. Army Corps of Engineers, especially CERL (Construction Engineering Research Lab). Since 1997 it is maintained by "The GRASS Research Group" at Baylor University, Waco (Texas), U.S.A. Several people all over the world are also developing modules - discussions and exchanges are done mainly through Internet via email, WWW-servers and two newsgroups. Even extramodules are offered in the Internet, also the sources for the entire package in ftp-servers. They can be copied freely and are well documented. The GRASS package can be extended through selfwritten modules using the GRASS programming library (C language). Several institutes have produced such modules - for erosion predicting, watershed modelling, image classification, chemical solution transport etc.
The GRASS sources can be compiled on all unix-platforms like SUN Solaris, SunOS, HP, SCO, SGI Indy, PC’s running LinuX, DEC Alpha etc. The needed capacity is generally at least 150 MB. For some platforms precompiled binaries are available in Internet.
Of special interest in the last years is the free UNIX-platform on PC’s, called "Linux". It is a full system with XWindows graphical user interface. If you want to use GRASS on your PC (486 or better), you can install this public domain unix operating system "Linux" on your PC. The Linux-system requires at least 120 MB (in an own partition beside DOS/Windows). The GRASS-binaries for Linux need only 40 MB. If you want to compile the package yourself some 150MB more space are necessary to store the sources. A three button mouse is also required.
The GRASS-binaries in all other unix-platforms need more space than in Linux, about 150MB. The GRASS source code can be compiled with the C compiler, some modules are written in Fortran 77 (Linux also offers free compilers, they are also available for other UNIX-platforms).
1.2 General structure of GRASS
The structure of the package can be described as followed:
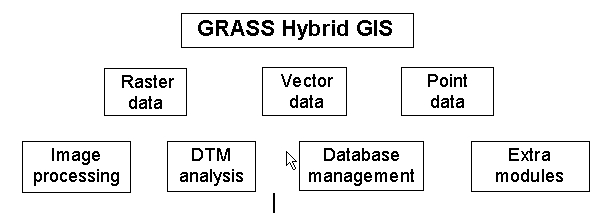
Conversion between all dataformats are possible. The user can also overlay layers in different formats. Direct overlays in the same format can be made with point data resp. vector data. To overlay raster data several modules are available like adding, multiplying etc. In raster image processing color composite generating is there. Data can also be overlayed with weights. The data may be displayed in the graphical output window or stored either in imageformats or postscript maps.
|
Main topic |
Modules |
|
Raster analysis |
Automatic rasterline and area to vector conversion |
|
Buffering of line structures |
|
|
Cell and profile dataquery |
|
|
Colortable modifications |
|
|
Conversion to vector and point data format |
|
|
Correlation / covariance analysis |
|
|
Expert system analysis |
|
|
Interpolation for missing values |
|
|
Neighbourhood matrix analysis |
|
|
Raster overlay with or without weight |
|
|
Reclassification of cell labels |
|
|
Resampling (resolution) |
|
|
Rescaling of cell values |
|
|
Statistical cell analysis |
|
|
Surface generation from vector lines |
|
|
Vector analysis |
Contour generation from rastersurface |
|
Conversion to raster and point data format |
|
|
Digitizing with board or on screen (scanned rasterimage) with mouse |
|
|
Reclassification of vector labels |
|
|
Superpositioning of vector layers |
|
|
Point data analysis |
Delaunay triangulation |
|
Surface interpolation from spot heights |
|
|
Thiessen polygons |
|
|
Topographic analysis (curvature, slope, aspect) |
|
|
Image processing |
Canonial component analysis (CCA) |
|
Color composite generation |
|
|
Edge detection |
|
|
Frequency filtering (Fourier, convolution matrices) |
|
|
Fourier and inverse fourier transformation |
|
|
Histogram stretching |
|
|
IHS transformation to RGB |
|
|
Image rectification (affine and polynomial transformations on raster and vector targets) |
|
|
Ortho photo rectification |
|
|
Principal component analysis (PCA) |
|
|
Radiometric corrections (Fourier) |
|
|
Resampling |
|
|
Resolution enhancement (with RGB/IHS) |
|
|
RGB to IHS transformation |
|
|
Texture oriented classification (sequential maximum a posteriori classification) |
|
|
Shape detection |
|
|
Supervised classification (training areas, maximum likelyhood classification) |
|
|
Unsupervised classification (minimum distance clustering, maximum likelyhood classification) |
|
|
DTM-Analysis |
Contour generation |
|
Cost / path analysis |
|
|
Slope / aspect analysis |
|
|
Surface generation from spot heigths or contours |
|
|
Screen displaying |
3D surfaces |
|
Color assignments |
|
|
Histogram presentation |
|
|
Map overlay |
|
|
Point data maps |
|
|
Raster maps |
|
|
Vector maps |
|
|
Zoom / unzoom -function |
|
|
Map creation |
PPM-image maps |
|
Postscript maps |
|
|
Extra modules |
Database interfaces (Informix, Ingres, Postgres) |
|
(several other modules are available) |
Erosion modelling (AGNPS, ANSWERS) |
|
Landscape structure analysis |
|
|
Solution transport |
|
|
Watershed analysis |
One of the main questions in a GIS is how to integrate the external data sources. This chapter describes the dataformats which can be imported into GRASS. If the data have a different format which is not supported by GRASS, then they can mostly be converted before importing. Several external programs for format conversions are available (especially for image formats). A famous program is "xv" which can be found in internet and compiled on any unix-platform. Another package are the "netpbm-tools" which are also stored in the Internet.
GRASS accepts a variety of raster dataformats - most of them are equal to standard image formats.
These formats can be imported:
1. ASCII (X Y Z values, z value range from 2 E7 to -2 E7)
2. TIFF (8bit, cell value range 0..255, with none, LZW or PackBits compression)
3. GIF (8bit, cell value range 0..255, GIF87-encoding)
4. PPM (24bit, cell value range 0..16.7 mill.)
5. SUN Raster (8bit, cell value range 0..255)
6. NHAP (aerial photo imagery format)
7. BIL/BSQ (satellite imagery format)
8. LANDSAT TM, MSS (satellite imagery format)
9. SPOT (satellite imagery format)
Supported export formats are:
1. ASCII (X Y Z values, z value range from 2 E7 to -2 E7)
2. PPM (24bit, cell value range 24 bit)
3. PPM/3 (3*8bit, the image is splitted into R, G and B band)
4. TGA (24bit, cell value range 0..16.7 mill.)
5. TIFF (8bit, cell value range 0..255, with none, LZW or PackBits compression)
One disadvantage in GRASS version 4.1 and 4.2 is the limitation of raster cell values to be only whole numbers. If the user wants to work in raster with floating point numbers, he/she should multiply the cell values with 10, 100 etc. and divide later the final resulting rasterlayer by this number. Another way is to store the floating point values in the category file (see chapter VI.11 in "raster data management" for further explanations).
GRASS will get floating point capabilities soon in version 5.0, which is planned in summer 1998. It will be made available through GRASS Research Group. In point- and vectorformat are no limitations.
Please be aware of the fact that the size of rastercells is at least one (of your defined units). A rastercell size cannot have for example the resolution 0.1 (m, inch etc.). Normally the ground resolution is defined - satellite images have for example 30m (LANDSAT TM). So the resolution of one rastercell is defined as 30m in width and height (x and y extend). If you want to work in resolutions less than one meter, define your resolution in centimeter etc..
GRASS vectors can be vectorpoints (not to be mixed with the GRASS point format) as well as line or polygon data. The exchange of these information is possible in several ways:
The accepted import formats for vector data are:
1. ASCII vectorformat
2. ARC/INFO (.arc, .line, .point and .textlabel files)
3. DXF
4. DXF3D
5. DLG (U.S. digital line graph format: optional format 3)
6. IDRISI
7. TIGER
To export vector data the following formats can be used:
1. ASCII vectorformat
2. ARC/INFO
3. DXF
4. IDRISI
5. MOSS
Point data, called "sites" in GRASS, are the third possibility of an dataformat. They are similar to vectorpoint data but stored internally in own tables. Conversion between vectorpoints and sites is possible. Point data may be spot heights or results of local measurements.
The point data can be imported in
- ASCII format (X Y Z values and a single word description for this point, delimiter may be
space, point or comma)
The Export is also possible in
- ASCII format (X Y Z values and a single word description for this point, delimiter may be
space, point or comma)
For map output GRASS offers a module for postscript maps and also maps in the image format PPM. Additionally there is an external mapcreation program "xmapgen". Both mapcreation programs are not very powerful, the definitions of the map layout have to be stored in a text file. Then the map is calculated by the GRASS module and the final result can be displayed with an imagery or postscript display program. So the user might be interested in using other external programs. For this purpose the layers can be exported and then imported into this external program.
1.1 Projections and coordinate systems
The first step in working with a GIS is the definition of the database. In a GIS the information are stored with coordinates - therefore a coordinate system has to be specified. The system which is to choose depends on the purpose - for raw satellite images mostly simple x-y coordinates without units are taken, for "real" geographical data the geographic system (latitude, longitude) or a geodaetic projection like UTM or Gauss-Krueger. To optimize the projection from the geoid into a flat map the earth is approximated by an ellipsoid. GRASS offers on the one hand predefined projections as well as the geographic system and on the other hand the possibility to define a geodetic projection yourself. If this feature is used, the projection type, the ellipsoid and the datum for referencing the ellipsoid has to be specified. The supported ellipsoids in GRASS are: airy, australian, bessel, clark66, clark80, everest, grs67, grs80, hayford, hough, iau76, international, krassovsky, merit, mercury, modified airy, modified everest, modified merc, new international, SEasia, sphere, walbeck, wgs66, wgs72 and wgs84. Defining an own projection can be done in the following systems: latitude-longitude, UTM, State Plane, Albers equal area, Lambert conformal conic, Mercator and in transverse Mercator (for Gauss-Krueger).
1.2 What information are necessary ?
To specify your database the following information are necessary:
· the coordinate system for the database (plain or with projection and ellipsoid)
· the minimum and maximum coordinates of the area of interest
· the ground resolution
Before the database can be created all these information have to be collected.
The first step in now to start the GRASS-package (the $ symbol is the unix prompt):
$ grass
In the first screen the name of the "location", the "mapset" and the GRASS-directory has to be specified. This GRASS directory must be created before using GRASS for the first time. Here are all data stored. Normally the user does not have to modify any GRASS-datafiles in this directory.
The "location" is the entire area. The "mapset" is the active used part which can be smaller or as big
as the location. Several mapsets can be defined within one location. The first screen displays:
After entering the names and the database-directory this screen can be left with
<ESC>. Now the required definitions for the project area (location) are requested. The names given in the first screen are always stored for the next start of GRASS.
Now the coordinate system has to be chosen. As descibed above, the plain xy-system, several projections and the geographical latitude-longitude system are available.
The procedure in each particular projection is as following:
2.2 The Projections and coordinate systems
(0) X-Y-system
Choosing the xy-system (for imagery and other unreferenced raster data) GRASS expects to
· enter a one line description and
· to define the default region (= location).
The origin is in the lower left corner. The south and east value will be calculated from the image size. The resolution is always 1 in xy-system.
Example:
Image size: 800 * 512
Definition of the default region:
DEFINE THE DEFAULT REGION
======= DEFAULT REGION ========
| NORTH EDGE: 512 |
| |
WEST EDGE | | EAST EDGE
0 | | 800
| SOUTH EDGE: 0 |
===============================
PROJECTION: 0 (xy) ZONE: 0
GRID RESOLUTION
East-West: 1_________
North-South: 1_________
AFTER COMPLETING ALL ANSWERS, HIT <ESC> TO CONTINUE
(OR <Ctrl-C> TO CANCEL)
(1) UTM
Using the UTM-projection the following information are required:
· enter zone (e.g. 30)
· enter a one line description
· define the default region (= location): specify your UTM-coordinates
· choose the ellipsoid (see list above or enter "list" here).
(2) State Plane
This projection is an U.S. specific projection.
(3) Latitude-Longitude
This system is a geographical reference. Here the user has to enter
· enter a one line description
· define the default region (= location): specify the latitude and longitude in decimal degrees.
Example: Madras (all values have to be entered in decimal degrees)
DEFINE THE DEFAULT REGION
======= DEFAULT REGION ========
| NORTH EDGE: 13.0 |
| |
WEST EDGE | | EAST EDGE
80.1 | | 81.0
| SOUTH EDGE: 12.0 |
===============================
PROJECTION: 3(Lat/Long) ZONE: 0
GRID RESOLUTION
East-West: 0.01______
North-South: 0.01______
AFTER COMPLETING ALL ANSWERS, HIT <ESC> TO CONTINUE
(OR <Ctrl-C> TO CANCEL)
Result (in degrees, minutes, seconds):
projection: 3 (Latitude-Longitude)
zone: 0
north: 13N
south: 12N
east: 81E
west: 80E
e-w res: 0:00:36
n-s res: 0:00:36
total rows: 100
total cols: 100
total cells: 10,000
(99) Other projection - own definitions
With this option an own projection can be defined. For example the Gauss-Krueger-projection, which is very common in Europe, can be specified. It is a transverse mercator projection. What are the steps?
First "Other" (99) will be chosen in the projection menu. Then you
· enter a one line description
· define the default region (= location): the coordinates depending on the projection
· specify the projection name
· choose the ellipsoid (see list above or enter "list")
· specify the datum (reference of the ellipsoid)
· enter scaling and units.
Here is an example for the region definition screen (with Gauss-Krueger coordinates):
After the definition of the location the first screen appears again. The names and the GRASS-database directory may be unchanged - the screen has to be left again with
<ESC>. Now the mapset is created. The coordinates for your mapset will be set to the location coordinates in the beginning. Further on they can be changed manually or by using the mouse-zoom-module.The idea of defining several mapsets in one location is to have a clear datastructure. For example several quarters in a city can be managed. Normally the data of other mapsets in a particular location are unvisible to the active mapset, but they can be accessed through a special access-module. But it is untypically for a GIS, that data for different areas are identical.
With returning to the standard unix-prompt the GRASS-package is activated. Beside all unix-programs the GRASS-modules (more than 300) are useable now. Very important is to leave the package properly after finishing work. Otherwise the datastructure may be damaged. To leave the GRASS software enter the command:
$ exit
GRASS asks now for the saving of the data. The default is "yes". Here (and also in the package) single files can be deleted.
III. General file management commands
Before starting with single command-descriptions the general command structure shall be explained. The command-structure is very easy to understand - the topic of the command determines the first letter of the GRASS module followed by a dot. The next word defines the subtopic, some command also have a subsubtopic. The following groups can be found:
|
d.* |
- display commands for graphical screen output |
|
g.* |
- general file management commands |
|
i.* |
- image processing commands |
|
r.* |
- raster processing commands |
|
v.* |
- vector processing commands |
|
s.* |
- site processing commands (point data) |
|
m.* |
- miscellaneous commands |
|
p.* / ps.* |
- map creation commands |
|
... |
- unix scripts |
Some examples for GRASS-commands: The name for displaying a image histogram becomes "d.histogram", the satellite image rectification module is named as "i.rectify", renaming a file can be done with "g.rename". For a single line description of more than 250 commands, please refer the Appendix 3: "GRASS Command overview" in this paper.
GRASS stores its data in an own directory, called the "GRASS database". Vector data are stored in a specific binary format, site data (point data) in ASCII lists, raster (imagery) data also have a specific compressed binary format. Access to the "outside" world is given through several modules (refer chapter I.2 in this document for the exchange formats). Normally the user should not affect files in the GRASS database. The management is done by the modules themself, to say, the files are physically "invisible" for the user.
2. Help system and file commands
Online help is available through display- and printable postscript-documents, local WWW-pages or with using the GRASS module:
$ g.manual
If this command is directly entered, a command list can be shown. The other way is to give a GRASS command name as parameter:
$ g.manual i.rectify
If there is no manual-page available, a short description can be displayed with the "help" parameter for the module:
$ i.rectify help
A better overview is given by the WWW-manpages. Free programs like
mosaic, netscape or lynx can display them. These programs can be easily installed in unix. Open in such a program the main WWW-file "grass.html" as local file. This file can be found in the grass directory, subdirectory ./html (for example /usr/local/grass4.2/html/grass.html).
The printable documentation (postscript files and textfiles) is stored in the
./documentation directory (e.g. /usr/local/grass4.2/documentation/). For previewing and printing of postscript files use$ ghostview
and
$ gs
All commands can be used interactively (enter only the command name), then the module asks for filenames etc. Or all required information can be given as parameter, the "help" parameter shows the syntax. This is important for using GRASS commands in unix-scripts.
Examples:
|
Purpose |
Command |
|
Copying of files |
$ g.copy |
|
Deleting |
$ g.remove |
|
List filenames |
$ g.list <datatype> |
|
Renaming |
$ g.rename |
3. Deleting of mapsets and locations
Sometimes it might be interesting to remove complete mapsets or even locations. A mapset can be deleted after giving the "exit" command to leave GRASS. Then the user is asked for removing single files or the entire mapset.
Locations can only be deleted outside of GRASS. Leave GRASS and use the unix "
rm" command (or "rm -rf" to avoid to be questioned for every file).Please use for file-deleting in the GRASS database only the "
g.remove" command, otherwise the database structure may be disturbed.
1. Introduction to point data management
In environmental studies are very often data only for particular stations or locations available. That could be for example spot heights (elevation data), taken from a toposheet, drill hole data or measured outside or rainfall data, measured by a climatic station. Several modules are offered by GRASS to analyse and convert these data. Point data are called "sites" in GRASS. In opposite to vectorpoints stored in vectorformat this is an own format. Internally an ASCII table is created in the GRASS database.
1.1 Import and export of sites
The import and export can only be arranged in ASCII format. This format is the most common format and available in every spreadsheet program, if the original data are stored there. To create this file export the sites in your spreadsheet program in the following ASCII structure (the description is optionally, a header is not necessary):
Generally: Example:
X-value1 Y-value1 Z-value1 desc1 3570321 5776988 102.4 Adyar
X-value2 Y-value2 Z-value2 desc2 3571987.3 5776876.4 110 Airport
X-value3 Y-value3 Z-value3 desc3 3574987 5777987.5 132.2 TTKRoad
...
Are the point data not stored in a spreadsheet program use a simple textedior to create this ASCII-file. The first two values (x and y) may be the coordinates while the z-value is the measured value like a spot height. But don't give any units here. The x-value is the easting, the y-value the northing coordinate. Allowed field separators are "space", "tab" and a character to be specified. "Space" is the default field separator. A description can be any text without spaces (use for space the underline character) but is only optional. This description is called "label" in GRASS.
Import
The importmodule is started with:
$ s.in.ascii
As usual the module can be used either interactively (menudriven) or non-interactively by giving parameters. The best way is to store the import-ASCII-file in the active directory. First the "sites name" - the name for the file in the GRASS database has to be given, then the name of the ASCII file containing the sites (exportfile from worksheet etc.). After specifying the field separator the sites are imported. Are the data imported from a DOS-textfile, the DOS-ASCII has to be converted into unix-ASCII.
Use the program
$ dos2unix file.asc
for this conversion. If this program is not available in your unix ask your system administrator (it might have another name).
The user can check for the existance of the file in the GRASS database with:
$ g.list sites
This command gives a list of all available site-files in the active mapset.
Export
The export module works similar, but the output has to be redirected into a file (otherwise screen output is given):
$ s.out.ascii > file.asc
First the sitesname ("list" shows all sites in this mapset), then the field separator has to be given. After that the module allows to choose if either all sites in he location or only the sites of the mapset shall be exported. The last question concerns the output of site descriptions.
The resulting file is stored in the actual directory.
1.2 Display of the sites - displaycommands
What is the result? To get a graphical output on the screen first the graphical outputscreen has to be started. This window is called the "GRASS monitor". It is possible to use up to eight "monitors", named from "x0" to "x7".
Before starting the window be aware that neighter a graphical WWW-program nor the image-conversion program "xv" is running. Otherwise the management of the colortable will run into conflict. These critical programs can be started after starting the GRASS monitor. Use the module either interactively with
$ d.mon
or commandline driven:
$ d.mon start=x0 or short
$ d.mon x0
To close a GRASS monitor enter:
$ d.mon stop=x0
Now you want to display the sites. Sites can either be displayed with or without description (text label of each site):
$ d.sites
(each site is marked with a "x")$ d.site.labels (each label is given without "x")
If you don't want to change any options in these modules just give the site-filename as parameter.
Shall both be displayed ("x" and description) call both modules.
To erase the screen enter:
$ d.erase
This command is very important: after changing the active region (change of coordinates) always this command has to follow. Otherwise coordinates in the GRASS monitor are not set properly.
1.3 A few words about the GRASS monitors
The monitor is somehow sensitive: You should take care not to change the monitorsize with the mou se (use environment settings instead) and to close it after work with the "
d.mon stop=x0" command. If you only close the window from the unix-windowmenu GRASS cannot not realize that the window is closed. It will remain blocked for further use untill you enter GRASS again and stop it.Solution for the first problem: Was the size accidentially adjusted, the "
d.erase"- module might clean the monitor. Otherwise stop and start it again.Did you forget to close the monitor and have already left GRASS, start GRASS again and select the monitor by:
$ d.mon select=x0
Then you can stop it.
Every user in the unix-system can use one or more monitors. But one monitor can only be used once at time.
If you are switching the location to another location and both have different projections the monitor always has to be closed before exiting GRASS.
The new sites information layer can be taken for analysis purposes. First the analysis of elevation data like spot heights will be discussed, later of hydrological data. Finally the conversion into other dataformat is described.
The structure of the ASCII-importfile with spot heights is usually like this:
Easting Northing Heightvalue
Easting Northing Heightvalue
...
Due to the limitation of standard GRASS in the rasterformat, that only non-floatingpoint values as z-values (cell values) can be used, you should contemplate to convert these heights to whole numbers. This can be done by multiplying the heightvalues with powers of 10. This procedure is only of interest, if you want to convert these sites later into a rasterformat layer.
Unix offers a simple method to multiply the third column (or other columns) with 10, 100 or other numbers. Enter therefore:
$ awk '{print $1, $2, $3*10}' file.asc > newfile.asc
The first and the second column $1 and $2 are unchanged, the third column $3 is multiplied with 10 and everything stored in a new file. "Awk" is a standard unix program. Remaining decimals will be rounded in the conversion process to rasterformat.
The imported spot heights can be used for several topographical calculations:
· slope, aspect
· profile curvature (measured in the direction of steepest slope)
· tangential curvature (measured in the direction of a tangent to contour line)
· mean curvature
With an existing rastermap (rasterlayers are described later) it is possible to mask areas of interest and non-interest. The corresponding GRASS module is:
$ s.surf.tps
It stores the results in only in rasterlayers and interpolation is done during the calculations. These rastermaps are like surfacemaps.
In this module several options can be set for the calculation:
· dmin1: Set minimum distance between points. Default value is set to 0.5 grid cell size of the resolution defined during the definition of the database.
· zmult: Convert z-values using conversion factor. Default value is 1 (use this option instead of awk, if you like).
· tension: Set tension. Default value is 40, appropriate for smooth surfaces.
· smooth: Set smoothing parameter. Default value is 0, no smoothing is performed.
· segmax: Set max number of points per segment. Default value is 40.
· npmin: Set min number of points for interpolation. Default value is 150, for data with heterogeneous spatial distribution higher value is suggested.
But mostly you will accept the default values. The algorithm is spline interpolation with tension.
Display of the results
Because the interpolation results from sites are mostly in rasterformat, here a small outlook to the display of raster images is given. This topic is described more specific later on. Before displaying the results, the GRASS monitor (e.g. x0) has to be started as described above.
The command to display rasterimages is:
$ d.rast rasterfile
If you want to look up the filenames of the calculated rastermaps enter:
$ g.list rast
You see the raster layers calculated above and can display them using the "
d.rast" command. Now it might be interesting to have a closer look to some particular areas in the active mapset. The command for zooming in a rastermap is called:$ d.rast.zoom
After entering this command move the mouse to the GRASS monitor, click a desired corner of the area of interest. Moving the mouse opens a window, the right button accepts the chosen portion.
Unfortunately there is not yet any mouseoriented "zooming-out" tool. The "
d.rast.zoom" offers to unzoom to the last zoom step. Otherwise the mapset has to be set to its defaults (the maximum extent), after that portions can be zoomed again. To set the mapset to its default coordinates enter:$ g.region -d
$ d.erase
Always the "
d.erase" has to follow to transmit the new coordinates to the GRASS monitor. Otherwise you either see the same resolution and coordinates as before or nothing any more.The "
g.region" module allows, if used interactively, to change the coordinates manually.
2.2 Interpolation of surfaces from spot heights
GRASS offers three module for the surface interpolation. This surface is stored in the raster format. Important is to know the number of spot heights, otherwise the interpolation may not be done properly. So first you count the number of given points in the ASCII-file containing the spot heights. For this you use the standard unix command "wc" (word count, which also counts lines with -l option):
$ wc -l spotheights.asc
As result you get the number of given points. With this information the interpolation can be started:
$ s.surf.idw in=spotheights out=elevation npoints=<number>
The input is the sitesfilename (GRASS name, can be looked up with "
g.list sites"), <number> the above counted number of points and output the new rastersurface. With:$ d.rast elevation
you can display the DEM on the screen.
If you want to use the "Kriging" algorithm, call the module
$ s.surf.krig
This module is an example for external extensions of GRASS, it was developed by an U.S. university. Information about Kriging can be found in the help-pages of SURFER(TM). Without proper parameters it will not produce any results.
Then the above described module:
$ s.surf.tps
calculates a DEM from given spot heights (splines with tension algorithm).
2.3 Statistical report on sites files
To get some information on the point data reports can be prepared with the module:
$ s.menu
This module is menudriven - you can easily get some statistical information.
Triangulation
The standard GRASS distribution offers two modules for triangulation. In GRASS 4.2.1 two new packages can be found - the v.geom- and the s.geom-package, where some improvements and additions have been done. Here the description for the standard modules follows.
To perform a convex hull shape triangulation, the module
$ s.delaunay
can be used. The result is a vector layer which can be displayed with
$ d.vect
To get a list of all available vectorlayers, take the module
$ g.list vect
To calculate Thiessen polygons in hydrological analysis of precipitation data there are two posibilities. The result can be stored in two different formats, either in vector areas (polygons) or in raster format. The choice depends on the further work. The input file is as usual a ASCII-file (see above) with Easting and Northing coordinates of each climate station and the rainfall amount as third parameter.
The module
$ s.voronoi
creates Thiessen polygons in a vector layer.
To store the calculated polygons in the raster format, use
$ s.surf.idw in=rainfall out=thiessen npoints=1
Important is to set the number of interpolation points (npoints) to "1".
4. Conversions of sites to other formats
This type of conversion does not perform a interpolation. The sites are directly converted into the rasterlayer without correction of coordinates. The non-occupied cell values are "0".
$ s.to.rast
To move the points onto a given vectorgrid (create with "v.mkgrids") you can use the polishing-module:
$ s.medp
All sites are moved to the next vectornode of the vectorgrid. The result is a new sitesmap.
To convert sites into the vectorformat use the module:
$ s.to.vect
The sites are stored as vectorpoints.
The resulting vector layer can be displayed with
$ d.vect
1. Introduction to vector data management
The vector datatype is based on shape oriented structures. It describes only the outline of structures. These structures may be linear structures like roads or streams or areal structures like lakes or fields. For example: You have cadastral data like ground plans of houses, so you are not interested what happens inside these rectangles. The required datatype would be vector - only the outline of the shape is of interest. Because of this less capacity is needed to store the information in the computer: The starting and the ending coordinates, the type of the structure (line, polygon etc.) and the attribute.
ARC/INFO format
Mainly you will have two different data sources: Maps on paper or vectormaps already prepared in an other GIS. The first topic will be discussed later on, here first follows how to import vectorfiles. Because the ARC/INFO vectorformat is most common only this topic is discussed here. Many GIS offer an export into this format which can be read in into GRASS.
The following chapter explains how to export the data from ARC/INFO itself. There are slight differences between the PC-based version of ARC/INFO and the UNIX-based version.
1.1.1 Export out of PC-ARC/INFO
The coverage-type can be "polygon" or line - the "describe" command helps you in ARC/INFO to find out. The precision must be "single precision". To export the data, you open your coverage in ARC/INFO and use the "ungen" command :
arc> ungen <datatype> <coveragename> <exportfilename>
The data are organized in two different structures:
· the line data (with start and ending coordinates)
· the label points (point coordinates for the label of each line) - only in polygon coverages
· the labels itself
All information have to be stored into different files. So you first export the linevectors resp. the polygons from the coverage (either a) or b), it depends on your ARC/INFO coverage type):
a) Type: line coverage
Export the linevectors into the lines-vectorfile (example: coverage name might be topo12):
arc> ungen line topo12 topo12.lin
b) Type: polygon coverage
Export the polygonsvectors into the polygon-vectorfile (example: coverage name might be topo12):
arc> ungen poly topo12 topo12.pol
Now the labelpoint data will follow (for polygon type):
arc> ungen points topo12 topo12.lab
The linked attributes have to be exported from the database used in ARC/INFO. Normally this is dBase in the PC-version. Store this table (pat.dbf) into a ASCII-textfile. The header has not to be removed from this textlabel file. Then you can continue with the import into GRASS.
1.1.2 Export out of UNIX-ARC/INFO
The export from points, lines and polygons is identical to the PC version. Only the database management differs, UNIX-ARC/INFO uses the INFO database.
After the export of the line or polygon coverage you have to start the INFO database program
within ARC/INFO:
arc> info
Now caps-letters are required.
The required username is "arc". Please give the following commands now:
ENTER COMMAND> SELECT <COVERAGE>.PAT
ENTER COMMAND> OUTPUT ../<coverage>.txt
ENTER COMMAND> LIST PRINT
ENTER COMMAND> Q STOP
Example:
ENTER COMMAND> SELECT TOPO12.PAT
ENTER COMMAND> OUTPUT ../topo12.txt
ENTER COMMAND> LIST PRINT
ENTER COMMAND> Q STOP
The result is the ASCII-file topo12.txt. Its structure is:
$RECNO AREA PERIMETER TOPO12# TOPO12-ID TOPO12-NAME
1 -2.30228E+0 19399.848 1 0 forest
2 81079.875 1678.826 2 1 wetland
3 955952.500 10229.637 3 2 lake
Now you can leave ARC/INFO. Then you can continue with the import into GRASS.
The three exported files have to be stored into the GRASS database manually (one of the few moments, you are allowed to do that...). Go into the directory
grass_data which exists in your home directory:$ cd grass_data
Then you change into the subdirectory with the name of your location, and within this directory again into your mapset (again a subdirectory). Is your location for example chennai and your mapset adyar that would be
$ cd chennai
$ cd adyar
In this directory there you create a direcory
arc.$ mkdir arc
Into this directory you copy ("
cp"-command) all three export-files.Then you can go back to you home directory.
Now the import can begin:
$ v.in.arc
|
Question: |
Line-coverage in ARC |
polygon-coverage in ARC |
|
The first parameter is the name for the new vectorlayer in GRASS |
topo12 |
topo12 |
|
The next parameter is the coverage "type" in ARC/INFO: |
line |
polygon |
|
With polygon-type it is asked for a neatline: |
------------ (not required) |
no |
|
The next parameter is the "linevector"-file from ARC/INFO: |
topo12.lin |
topo12.pol |
|
Then the "labelpoints" will be specified: |
------------ (not required) |
topo12.lab (if not available, press return) |
|
The "label texts" are the attributes (only read in when labelpoints there): |
topo12.txt |
topo12.txt |
If labelpoints are there the import module shows the beginning of this label file (the first three lines).
It may (!) look like this (so be careful), depending on the ARC-version:
$RECNO AREA PERIMETER TOPO12# TOPO12-ID TOPO12-NAME
1 -2.30228E+0 19399.848 1 0 forest
2 81079.875 1678.826 2 1 wetland
3 955952.500 10229.637 3 2 lake
The import-module asks now for the number of the ID-column. That will be the column with continous numbers (here: 4). The CAT-column (category value) is the next following column-number (5), the ATTRIBUTE the number of first text column.
The import will be completed then. After this module the creation of the support files have to follow:
$ v.support map=topo12
Here you specify the new GRASS vectorlayer as filename. The idea of this module is to reorganize the vectorlayer properly.
After the starting of a GRASS monitor the imported layer can be seen with
$ d.vect topo12
GRASS also supports the export into the ARC/INFO-format. You start the module
$ v.out.arc
It asks for the coverage type (line or polygon), the GRASS vectorlayer and the prefix to be given for the file extensions (.lin.,.lab,.txt). You can specify here the same name like the GRASS vectorlayer.
You should export both coverage types from the same file, only in this case line as well as polygon structures are exported. So this module has to be run twice.
Attention: The output is not stored in the home directory, but again (like above) in the GRASS database. Change the directory to find the files:
$ cd grass_data/<location>/<mapset>/arc/
Example:
$ cd grass_data/chennai/adyar/arc/
For the use of GRASS-vectorlayers in CARIS it is only possible to export the files into the DXF-format because CARIS does not accept ARC/INFO-format. First the vectorlayer has to be cleaned and then converted from the internal binary format into the internal ASCII format. Choose the module:
$ v.support map=vectorlayer
$ v.out.ascii
It asks for the existing binary vectorlayer and then for the name for the new ASCII-layer. Give the same name. To export the file use:
$ v.out.dxf
The module asks for the ASCII-layer name and the new DXF-name. The file is created and can be taken from the home-directory.
The import of DXF into GRASS is actually a problem with newer software because of changes in the DXF-command structure. But the module could be updated because the source code is available.
Digitizing is used to transfer information from papermaps like toposheets or thematic maps into a GIS. GRASS provides support for digitizer board as well as mouse digitizing on the screen (in the GRASS monitor). The second way is less costly, "only" a scanner is required to scan the map on paper. Then this scanned image can be imported as rasterlayer (xy-system) and rectified into the location with geographic coodinates or projection. For this purpose you will use the
i.rectify-command, assign geographic coordinates to the four corners of the image and then transform it (affine transformation). The result is a geocoded map in the location. This is discussed later on in chapter VII "Image Processing". This scanned map can be used as "backdrop-map" in the digitizing module. With the mouse on-screen digitizing (with zooming and panning) is possible.
The configuration of a digitizing board is somehow difficult (as usual in GIS): The configuration file has to be set properly and then the driver to be compiled (C-compiler). There are several precompiled drivers available. But here only the more easy way of using scanned maps will be discussed.
The scanned map has to be stored in a standard image format. The best choice is, because the map will be in colors or greyscale (both a 8bit format in computers), to store it in in the following way:
1. colorimage: GIF (8bit = GIF87), SUN Raster (8bit), TIFF (8bit with none, LZW or Packbit compression), PPM (24bit)
2. greyscale image: TIFF (8bit with none, LZW or Packbit compression) or GIF (8bit = GIF87)
Every scanner software will support one of these image formats. The file has to be stored in the home directory on the unix machine. Because this image being without georeference, it has to be transformed. See the chapter VII "Image processing" later on, in what way the xy-location has to be defined and the affine transformation to be performed.
It shall be suggested now that the image is already in a database with projection (UTM, Lat./Long. etc.). With
$ d.rast mapname
you can, as usual, display the map in the GRASS monitor.
Now the digitizing module can be started:
$ v.digit
Please do not change the size of the terminal window below the size of 80x25 (simply take the original size). Otherwise the menusystem from v.digit cannot be opened.
The first choice is the digitizer, if you want to digitize using the mouse, choose "none". Then the module asks for the name of a vectormap layer (existing or new name).
After that a new screen opens:
Here you can fill out the first 5 lines (not necessary). Important is to set the "Map's Scale" properly. For the mouse digitizer choose:
Map's scale: 1:1
If you are using a digitizing board, specify here the map scale of the papermap. The coordinates are taken from the region-setting. If you are using a digitizing board, the referencing follows. After confirmation of the next message the following screen, the digitizing menu, appears:
With pressing the first letters (In caps! Please look careful for these first letters) you enter the me-nues. In "Digitizing" you can digitize, "Edit" allows later on corrections, "Label" allows labeling (assigning attibutes to the vectors), also automated contour labeling. "Customize" has the menu to display the scanned map (choose "Backdrop cell map" and give the name of this map). "Zoom" is also important, you can magnify particular regions or use the "panning" function (that is shifting the middle of the area, useful to "move around" on the map). If a "Buttons choice" message appears, always the mouse has to be used in the GRASS monitor.
This module is self explaining and has a help function. If you leave "
v.digit" the map will be saved.For more explanation please refer the "GRASS tutorial: Digitizing" (see References).
3. Conversion of vector layers to other formats
Before using the conversion routines you should always use the "clean-up"-module:
$ v.support map=vectorlayer
3.1 Vector format to sites format
The conversion to the sites format (if you digitize spot heights with height information as label) is done with:
$ v.to.sites
3.2. Vector format to raster format
The conversion to rasterlines (from lines) and rasterareas (from polygons) is performed with:
$ v.to.rast
4. Overlay and splitting of vectorlayers
Several vectorlayer can be overlayed into one layer with the module:
$ v.patch
To split vectorlayers use the module
$ v.cutter
After this
$ v.spag -i
has to be run. This cleans unused nodes and double lines from the layer. Please refer the manpages (g.manual or WWW) for further information.
1. Introduction to raster data management
The use of raster data offers a nearly "never ending" variety of possibilities to calculate and derive maps. The input may be toposheets, thematic maps, aerial photos or satellite images. From these data cellwise or matrix-oriented calculations can be carried out to create new thematic maps. Later on in this training also image enhancement and the derivation of thematic maps from satellite images will be discussed. Very interesting is the feature of raster maps that the neighbourhood influence can be calculated: This allows to create transportation models (for soil erosion, water movement, pesticide movement etc.).
Two different facts have to be distinguished in raster data: On the one hand there are the cell values, the "z-values" with coordinates. On the other hand a colortable can be assigned to these cell-values depending on the users purpose. Both is possible: Either each cell has its own color for the display or color is assigned groupwise to the cell-values, they are "classified" in this case. About this you
should consider carefully, because a change in colortable does not affect the cell values. That is important for the calculations further on.
In scanned maps the colors of the original map determine the cell values, they are the same. But after the import into a GIS like GRASS the user can either assign other colors to the map without changing the cell values or change the colors of the map (without having a colortable) by changing the cell-values. In this case the system itself assigns the colors.
1.1 Import and export of rasterdata
To import raster data like scanned maps, satellite images, scanned airphotos etc. several import facilities are offered. The choice of the imageformat depends on the source you want to import.
1.1.1 Scanned maps (greyscale or color)
If you scan a map you have normally 256 colors or greyscales (both 8bit). The scanning software offers several image formats, you can choose here the GIF format (GIF87). GIF89 is not (yet) supported, because it is a 24bit format.
To import a scanned image, store it into the working directory. Because an image is not georeferenced there are two choices:
1. The images fits into the entire location or into a part of it.
2. The image has to be rectified.
We discuss only the first possibility here, the second will be discussed in the chaper "Image processing" later on.
For the first choice you must know the boundary coordinates of the image map and its ground resolution (for example 30m). Ground resolution means the extent each raster cell has. This can be calculated from the resolution chosen in the scanning process (dpi = dot per inch = lines per inch).
Example:
Scanner resolution: 300dpi. Scale of the map: 1:100.000
One centimeter is equal to 1000m.
300dpi = 300 lines per inch = 300 lines/2.54cm = 118.11 lines/cm
Per centimeter the ratio is: 1000m/118.11 lines = 8.47m/line
The raster cells are squares, therefore the cell resolution is 8.46m x 8.46 m.
Now you set the coordinates and the calculated resolution in GRASS:
$ g.region
The menue 1 allows: "Modify current region directly". Type in here the coordinates of the image and the ground resolution.
After leaving this module you must use
$ d.erase
to send the new coordinates to the GRASS monitor.
Then you can import the image:
$ r.in.gif
The first filename is the GIF-file stored in your working directory. The second name is the new rastername to be used in GRASS. Optional a title can be specified. For the "verbose" mode say "yes" - you get information about the progress of import or errormessages, if the imageformat does not fit.
For TIFF you type in:
$ r.in.tiff
The use is similar to GIF import.
After each import the module
$ r.support
has to be run interactively. Note: It is the similar to the situation when you import vectorlayers and use "
v.support" afterwards on this vectorlayer.
GRASS allows you now to adjust the rasterlayer coordinates, because it cannot guess the proper coordinates itself. So specify as filename the name of the just imported rasterlayer.
· "Edit the header?": y
· "Please enter the following information...": <ESC> (leave this page unchanged!)
· Now the coordinates of the image can be specified. Give here the boundary coordinates of your image as you have done in the
With <ESC> you leave this screen after entering the rasterimage coordinates properly.
· "Update the stats...": y
· "Edit category file": n (for the future: here you can change textlabels assigned to cell values)
· "Create/update colortable?": n
· "Edit the history file...?": n
The file is imported properly now. You can display the imported image with
$ d.rast
1.1.2 Rasterdata with range from 0 to +16.7 million (24bit)
The second possibility is to store data in PPM-image format. The procedure is the same as using the GIF import module. For PPM you use
$ r.in.ppm
If you want to import images from three channels (RGB), then you can convert each of them to 8bit-PPM and import it with
$ r.in.ppm3
All the image format conversions can be done with the program
$ xv
which belongs to the standard LinuX distribution (shareware, can be compiled on every unix-platform). You load the image (load button) and save it again, in the save menu you choose your image format.
1.1.3 ASCII-Data (positive and negative values)
If you have data with positive and negative range (e.g. bathymetric data) you cannot use image formats because they don't support negative values. This type of data can be imported in a ASCII table.
The structure is the following:
north: 4928000
south: 4914000
east: 609000
west: 590000
rows: 700
cols: 950
28 28 28 28 28 28 28 52 52 52 52 51 51 51 51 51 51 51 51 51 51 51 51 52 52
52 52 52 52 52 52 52 52 51 51 51 51 51 51 51 51 51 51 51 26 26 26 26 26 26
47 47 47 47 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 51 44 44
44 44 44 44 44 44 44 29 29 29 29 29 29 35 35 35 35 35 35 35 35 35 35 35 35
35 29 29 29 29 29 29 29 29 29 29 29 54 54 54 54 54 54 54 50 50 50 50 50 50
50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50 50
...
The header has to be added to your ASCII values. The number of columns and rown depends on the distance between the east-west and north-south boundaries and the resolution.
Then the rastercell values follow: Sorted from north to south and west to east. That means, the data have to be exported into the ASCII-format linewise from left to right and from the upper boundary down to the lower boundary. After each ending of the line, there is for the next line a jump back to the left boundary.
Set the region properly (resolution!) with
g.region. The module's name for ASCII import is:$ r.in.ascii
After the import
$ r.support
should follow to adjust the coordinates from the rasterlayer.
Then it can be displayed with
$ d.rast
For positive values up to 255 (export in 8bit format) you can use
$ r.out.tiff
After specifying the file names you can choose if you want the file compressed.
Also the export can be performed in the 24bit image formats PPM and TARGA (positive values up to 16.7 million):
$ r.out.ppm
and$ r.out.tga
or also splitted into the three basic colors (result are three RGB PPM-files):
$ r.out.ppm3
With the "xv" program these image formats can be changed to others, also to 8bit GIF.
If you have negative and positive cell values, the ASCII format can be used for exporting:
$ r.out.ascii > textfile.asc
Here the output has to be redirected into a file.
As described above, the colortable is assigned to the cell values. These values are also called "category values". After importing a rasterimage no colortable exists. This does not matter because the system assigns colors itself. But sometimes the user wants to create a own colortable. There are several possibilities:
2.1 Assign a standard colortable
The first possibility is to use:
$ r.support
Here you find the possibility to choose a standard colortable, which can be also changed later on.
To assign this table you can also use:
$ r.colors
2.2 Change the colortable interactively
The module
$ d.colors
has a menudriven system to assign to each cell value (=category value) RGB-values manually.
To get an idea about these values a table of standard colors for toposheets follows:
|
Symboltype |
R |
G |
B |
|
0 - black - no data |
0 |
0 |
0 |
|
1 - white color |
255 |
255 |
255 |
|
2 - Forest (green) |
184 |
240 |
153 |
|
3 - Waterbodies (light blue) |
192 |
230 |
255 |
|
4 - Contours (brown) |
179 |
102 |
26 |
|
5 - Digs (dark blue) |
0 |
51 |
255 |
|
6 - Ground plans of houses etc. (black) |
0 |
0 |
0 |
|
7 - Text (dark blue) |
0 |
0 |
255 |
|
8 - Trees (green) |
0 |
192 |
0 |
|
9 - (dark grey) |
192 |
192 |
192 |
|
10 - (light grey) |
217 |
217 |
217 |
The external program "
xv" offers a menu to choose colors in an image by mouse. Here you can open any image and try out other color combinations.
The assigned colortable will be displayed in the GRASS monitor with the module:
$ d.colortable
Several options allow to optimize the output.
3. Display of raster maps in different scales
Sometimes it is very helpful to get a closer look at a particular area. For this purpose the module:
$ d.rast.zoom
was created. It only works after displaying a raster map. After the start with the left button a corner for an opening window can be choosen (one or several times). Moving the mouse opens then this window. With right mouse button it can be accepted, automatically the rastermap will be redrawn.
Unfortunately nobody has programmed a unzoom (widen view) module yet. Therefore you can only set the view to the entire location and zoom in again. To set the coordinates to the maximum extent, give these three steps:
$ g.region -d
$ d.erase
$ d.rast rastermap
Please don't forget that the first command will also set the resolution to the standard value.
Or you set the resolution directly to the raster map:
$ g.region rast=rastermap
$ d.erase
$ d.rast rastermap
4. Query raster cell contents and display of profiles
To get information about the coordinates and the category value of each particular cell the mouse-driven module
$ d.what.rast
can be used. It only works after displaying a raster map. With the mouse you can click on a rastercell, the output is in the terminal window. Leave this module with the right button.
In the profile module you can choose a rastermap, and in this map a transect (line) from which a profile shall be drawn. The modules name is:
$ d.profile
The module is menu-driven and selfexplaining. The profile is drawn from the category values.
Other modules are r.transect and r.profile, they give raster values in ASCII format which can be redirected into a textfile.
5. Classification of raster maps
The classification of a map is a manipulation of its cell values. If you have a raster map with a huge amount of different information - a wide range - like in a elevation map, it is useful to classify this map. The cell values are assigned groupwise to certain classes which can be defined by the user. The module for the (re-)classification is
$ r.reclass
There are two possibilities in using this module, you can define each label manually by editing the categories in the tables or use the "prompt"-mode to categories values groupwise. This module modifies the raster map directly and does not affect the colortable. Changes in the colortable are
done with "
The first method is usable if you want to
· create binary masks only from one category value (it is set to 1, rest might be zero)
· modify only a few number of cell values manually
So you use the module without giving parameters.
The second method is usable if you want to
· classify the image with rules (classify groupwise)
So you use the module with giving the input and the output mapnames.
First method
If you start it without parameters, it asks for the source mapname and then for the new output map name. After that you are queried, if you want to preset the new values to 1 or 0.
A watershed example: Using the reclass-module interactively is helpful if you want to create a mask for the "
r.mask" module. First you digitize a watershed boundary in "v.digit", give here this polygon-area a value, convert it to raster format with "v.to.rast". Then you reclass the cell values of the entire raster map, so that to the watershed the value 1 is assigned and to all outside areas the value 0. The "r.mask" module puts this mask onto the location so that only the watershed area is used for calculations (minimize boundary calculation errors by this).After that the modification screen is opened, where you can assign the new cell values. With <ESC> you can go through the pages. The textlabels (category label) can be modified using the "
r.support" module.
Second method
If you specify the input and the output filenames
$ r.reclass input=sourcemap output=newmap
you get into the "rules definition mode", a prompt is displayed. Here you can assign groupwise a new category value (raster cell values) and a textlabel:
Example 1:
1 3 66 = 2 poor quality
23 44 67 = 3 better quality
This changes all category values 1,3,66 to 2 in the new map and gives the textlabel "poor quality".
Example 2:
1 thru 10 = 2 poor quality
10 thru 20 = 3 better quality
This changes groupwise: The calues from 1 to 10 become 2 in the new map etc.
The textlabel is only optional.
Example 3:
1 thru 10 12 thru 19 = 2 poor quality
11 = 3 better quality
25 = 2
Here two groups are assigned to "poor quality" and also 25. For 25 the assignment of the text label is not necessary because it is already defined, that 2 shall be "poor quality".
Attention:
The original map shall not be removed because the classified map stores only the classification-rules. If you delete the source then you cannot access the classified map any more. If you use
$ r.mapcalc <newfile> = <rescaled-map>
a physical new layer will be created.
In opposite to the classification of single cell values also the entire range can be changed. Example: You have a map with cell range from 1 to 1500 but you want to have it in the range from 444 to 888. Then you can linear rescale the raster map:
$ r.rescale
First you specify your source map then the new map. After that a menu appears. The min and max of the source raster map is displayed and also a preset given for the target map (1 to 255). You can modify these values for your purpose. Changing the parameters for the source map you can also rescale only a portion of the cell values, e.g. only set the values in the range from 800 to 900 to 80 to 90. This rescaling is done linear.
7. Managing label information in raster maps
To change the label information the module
$ r.support
is to be used. There is a menu to "Edit the category file for ...". Choose this option, leave the next screen with <ESC> and then you get the table of textlabels (called "category names" here). Here you can write what you want.
To display the assigned textlabels from a raster map, use
$ r.cats
This gives a table of the category values and their labels.
Two other modules to get information about the raster files are
$ r.describe and
$ r.info
8. Display of histograms and cell distributions
Usually the knowledge about histograms is important in satellite images. But this module can be used to visualize the distribution of raster cell values. The module is
$ d.histogram
Used interactively it displays the cell distribution in bar style, used with parameters also the "pie" style can be activated.
To get this distribution in a table, the module
$ r.stats
is there. It can either give a list of cell values (to get the range of the raster map), or count the number of cells for each particular cell value (option "-c"). With option "
-1" it displays every cell value, ordered from north to south linewise from west to east, with "-1g" also the coordinates of each cell are displayed (if you redirect the output into a file, you get a sites list of this raster map.). With "-l" also the labels are displayed.
If you digitize line structures and convert them to raster format, the module
$ r.buffer
allows you to give this line structure a width (e.g. the width of a road). Also noise disturbance etc. can be simulated.
To let points and areas "grow" like this, the module
$ r.grow
can be used. This adds one cell in each direction around the areas.
10. Example: Creating a potential soil erosion map
After looking at several modules separately today the first sequence of commands shall follow. Today the example of the introduction - the potential soil erosion map - will be calculated. This model is only very simple, using some basic information. The map showing the potential soil erosion for the entire area can be calculated from the
· slope map
· vegetation cover and
· erodibility (K-factor).
The slope map can be derived from the elevation model (DEM). It is a neighbourhood rastermatrix analysis, where for each particular cell the height of the neighbouring cells is surveyed and then according to the ground resolution the slope angle for this centered cell is calculated.
The vegetation cover has to be digitized and after that converted to the rasterformat. Before converting, the resolution should be set to the resolution of the DEM.
The erodibility can be derived from the soils map/ soils texture map. First the soils map must be digitized and converted to raster format (here also same resolution as DEM). Then a reclassification of the soil types has to be performed, assigning the specific K-factor (soil erodibility factor) to each soil type. This factor can be computed from the soil texture and structure of the upper layer (calcula-ted after USLE, if the % silt is less than 70%. Using texture: % sand, % silt, % clay; the % organic matter, structure and permeability).
Usually there are more factors which have to be used, but this is only a simple example.
The slope map can easily be calculated with the module
$ r.slope.aspect
First you specify the DEM, then names for the aspect and the slope file have to be given. As your wish you can calculate the slope in degree or in percent. Also it can be decided if zero elevations are true elevations (e.g. in costal zones) or are data errors (then they are ignored). Because very small slope degrees are calculated very unexact you can define a specific lower boundary here. In this example the slope shall be calculated in percent.
10.2 Generating the vegetation cover map
The vegetation cover has to be digitized from a map (v.digit) with area vectors (polygons). Each area has to get a label with its specific vegetation. Also the information of specific types of agriculture like irrigation or fallow land.
After digitizing this map layer has to be converted into the raster format with
$ v.to.rast
It is only converted properly if all vectors have a label.
10.3 Generation the erodibility map
This maplayer is a derivation of the soil texture (sand, silt, clay), the soil structure (course, medium, fine), the organic matter and the permeability. The calculation should be done following the Universal Soil Loss equation (USLE).
So first the soils map has to be digitized, then converted to raster format. Using the classification module
$ r.reclass
for each soil type the K factor can be assigned (using the USLE).
10.4 Classification of the input maps
Here an example for a simple soil erosion model follows. It depends only on the three input layers, they are classified and then overlayed with specific weights. It is important to "translate" the specific theme of each map to the erosion theme, that means to assign the grade of erosion to each input theme. The conditions in this non-scientific example might be:
Slope (in percent)
· slope: 0 -10% : 1 (mild erosion)
· slope: 11-20% : 2 (medium erosion)
· slope: > 20% : 3 (severe erosion)
Erodibility/K-factor
· erodibility : 0.1 - 0.2: 1 (mild erosion)
· erodibility : 0.21 - 0.35: 2 (medium erosion)
· erodibility : 0.36 - 0.5: 3 (severe erosion)
Vegetation cover
· vegcover: all forests types: 0 (no erosion)
· vegcover: rangeland: 1 (mild erosion)
· vegcover: irrigated: 2 (medium erosion)
· vegcover: disturbed: 3 (severe erosion)
To assign these classes use the module with "rules":
$ r.reclass
Examples:
Definition of slope classes (please enter also the description in braces):
$ r.reclass input=slope.percent output=slope.class
0 thru 10 = 1 ( slope < 10%)
11 thru 20 = 2 (10% < slope < 21%)
21 thru 1000 = 3 (slope > 20%)
end
Definition of K-factor classes:
$ r.reclass input=erode.index output=erode.class
0.1 thru 0.2 = 1 (0.1 thru 0.2 erodibility )
0.21 thru 0.35 = 2 (0.21 thru 0.35 erodibility )
0.36 thru 0.5 = 3 (0.36 thru 0.5 erodibility )
end
Definition of vegetation cover classes:
$ r.reclass input=vegcover output=veg.class
2 = 1 (1 rangeland)
1 = 2 (2 irrigated land)
6 = 3 (3 disturbed land)
3 4 5 = 0 (0 forest)
end
The final map "potential soil erosion" shall be calculated by a weighted overlay:
Slope: 40%
Erodibility: 30%
Vegcover: 30%
To perform this calculation use the module:
$ r.mapcalc
This module allows direct map calculations which are done cellwise.
The general formula to calculate the final map "potential soil erosion" from the input maps mapi with their specific weights wi (in percent) is:
n
finalmap = n/100 * å (wi * mapi)
i=1
To give this calculation in the module
r.mapcalc this formula is:potential.soilerosion = round(float(3 * ( 40 * slope.class + 30 * kfactor.class + 30 * veg.class)) / 100)
If you use r.mapcalc you should always give division as late as possible to avoid rounding errors. Please remember - in this actual version of GRASS only whole numbers in raster format are supported. Also it is recommended to give "blank spaces" before and after a an arithmetical symbol.
10.5. Validation of the result
Very important is the validation of the map. GRASS offers a very helpful tool:
$ r.cross
This module calculates the cross product of given raster maps. That means the input maps are compared with the output maps. In this case the input maps are the slope, the erodibility and the vegetation cover. It is compared with the potential soil erosion map. So all these maps are the input for this module, the output is a new raster layer.
To perform this calculation give:
$ r.cross input=potential.soilerosion,slope.class,erode.class,veg.class output=potential.soilerosion.cross
or use it interactively (after the last input map give one "return" to switch to the query of the output map). The resulting output can be displayed. The module
$ r.cats
describes the correlations between the input and the output map. Also you can use
$ d.what.rast
to query raster cells directly with mouse.
If this validation map is highly disturbed normally an error has occured in the calculation-formula. Verifying the "r.cats"-table you will find the error. The "normal result" will show small areas which are called "smallest geometries" of the overlayed input maps. If these input maps don't correlate, these "smallest geometries" will only have the size of a few raster cells.
11. Overview of the functions in r.mapcalc
This module comes along with an external description, in this chapter only the main functions are described. You can use the the module interactively or with the formula as parameter:
$ r.mapcalc
$ r.mapcalc result = expression
Operators and order of precedence
The following operators are supported:
Operator Meaning Type Precedence
--------------------------------------------------------------------
% modulus (remainder upon division) Arithmetic 4
/ division Arithmetic 4
* multiplication Arithmetic 4
+ addition Arithmetic 3
- subtraction Arithmetic 3
== equal Logical 2
!= not equal Logical 2
> greater than Logical 2
>= greater than or equal Logical 2
< less than Logical 2
<= less than or equal Logical 2
&& and Logical 1
|| or Logical 1
The operators are applied from left to right, with those of higher precedence applied before those with lower precedence. Division by 0 and modulus by 0 are acceptable and give a 0 result. The logical operators give a 1 result if the comparison is true, 0 otherwise.
Rules for the map names
To avoid problems the map names used in r.mapcalc should always contain a letter. Otherwise the map names have to be quoted to avoid, that they are interpreted as numbers.
Example:
mapcalc> newmap = 123 + 3
creates a map with only the value 126. In opposite to that
mapcalc> newmap = "123" + 3
adds the value 3 to the content of the map layer "123". That can be any result here.
If you give map names containing letters, confusion can be avoided.
The neighbourhood modifier
Maps and images are data base files stored in raster format, i.e., two-dimensional matrices of integer values. In r.mapcalc, maps may be followed by a neighborhood modifier that specifies a relative offset from the current cell being evaluated. The format is map[r,c], where r is the row offset and c is the column offset. For example, map[1,2] refers to the cell one row below and two columns to the right of the current cell, map[-2,-1] refers to the cell two rows above and one column to the left of the current cell, and map[0,1] refers to the cell one column to the right of the current cell. This syntax permits the development of neighborhood-type filters within a single map or across multiple maps (cited after r.mapcalc manpage).
Raster map layer values from the category textlabel file
If you want to use the cell labels (category labels) instead of the cell values (category values), for example to store floating point numbers in a raster cell then you simply specify the @ operator for this particular map.
Example: You have a soils pH-value map with the following labels:
cat label
------------------
0 no data
1 1.4
2 2.4
3 3.5 etc.
Then the expression:
mapcalc> result = @soils.ph * 10
would produce a result with category values 0, 14, 24, 35, 58, 72, 88 and 94.
If you want to store this also as category labels in the resulting map specify
mapcalc> @result = @soils.pH * 10
Functions
The functions currently supported are listed in the table below. The type of the result is indicated in the last column. F means that the functions always results in a floating point value, I means that the function gives an integer result, and * indicates that the result is float if any of the arguments to the function are floating point values and integer if all arguments are integer.
function description type
-----------------------------------------------------------------------
abs(x) return absolute value of x *
atan(x) inverse tangent of x (result is in degrees) F
cos(x) cosine of x (x is in degrees) F
eval([x,y,...,]z) evaluate values of listed expr, pass results to z
exp(x) exponential function of x F
exp(x,y) x to the power y F
float(x) convert x to floating point F
if decision options: *
if(x) 1 if x not zero, 0 otherwise
if(x,a) a if x not zero, 0 otherwise
if(x,a,b) a if x not zero, b otherwise
if(x,a,b,c) a if x > 0, b if x is zero, c if x < 0
int(x) convert x to integer [ truncates ] I
log(x) natural log of x F
log(x,b) log of x base b F
max(x,y[,z...]) largest value of those listed *
median(x,y[,z...]) median value of those listed *
min(x,y[,z...]) smallest value of those listed *
round(x) round x to nearest integer I
sin(x) sine of x (x is in degrees) F
sqrt(x) square root of x F
tan(x) tangent of x (x is in degrees) F
Floating point values in the expression
Floating point values in the expression are handled in a special way. With arithmetic and logical operators, if either operand is float, the other is converted to float and the result of the operation is float. This means, in particular that division of integers results in a (truncated) integer, while division of floats results in an accurate floating point value. With functions of type * (see table above), the result is float if any argument is float, integer otherwise.
However, GRASS raster map layers can only store integer values. If the final value of the expression is a floating point value, this value is rounded to the nearest integer before storing it in the result raster map layer (cited after r.mapcalc manpage).
Examples for if-constructions: (x is a mapname, all other letters can be numbers or maps.
The operation is done on each particular cell value of the map(s))
1. if x = a then b in r.mapcalc: result = if((x - a),c,b)
else c
2. if x <> a then b in r.mapcalc: result = if((x - a),b,c)
else c
3. if x >= a then b in r.mapcalc: result = if((x - a),b,b,c) (the second b is for x=a)
else c
4. if a <= x <= b then c in r.mapcalc: result = if((((a <= x) && (x <= b)) - a),c,c,d)
else d
12. Overview of GRASS capabilities in watershed modelling
The watershed modelling or nowadays "watershed management" deals mainly with the distribution, storage and usage of water (mostly rainfall water) in a watershed. To limit the area to a watershed minimizes the number of influencing factors.
The following pages give a short overview about the modules available in GRASS. Several other modules can also be used for this purpose - even the satellite image processing for creating landuse maps etc. is not included in this paper.
12.2 Watershed calculation and creation
The following descriptions are mainly taken from the GRASS manual pages and partly modified..
v.digit - GRASS digitizing module.
This module allows to digitize toposheets. For watershed analysis it will be used to digitize the contourlines, from which a DEM surface will be generated.
Either a digitizing board can be connected or scanned imported maps can be digitized on screen using the mouse. There is an extra manual available for this module.
v.in.arc - Import ARC/INFO vector layers into GRASS
v.in.arc allows to import ARC/INFO vector point, line and polygon features with their associated attributes (labels) into GRASS.
s.in.ascii - Import point data into GRASS
To import spot heights and climate data like rainfall this module can be used. The values have to be stored in an ASCII file with their easting and northing coordinates and the value (spot height, rainfall etc.).
r.in.gif, r.in.tiff, r.in.ppm - Import raster images into GRASS
These modules allow to read in 8bit and 24bit raster images like scanned maps, already prepared spatial data in raster format etc. Generally only positive values are supported.
r.in.ascii - Import raster values into GRASS
In opposite to the image format also negative values can be imported. The cell values have to be stored in an ASCII file with a header (containing the boundary coordinates, no. of rows and colums)
r.support - Setting raster layer parameters
With r.support the coordinates can be set to an imported raster image. Also the editing of the text labels to cell values can be done.
12.2.2 Calculating and setting the watershed boundaries
r.watershed - Watershed basin analysis program.
The module r.watershed generates a set of maps indicating:
1) the location of watershed basins, and
2) the LS and S factors of the Revised Universal Soil Loss Equation (RUSLE).
The Input map is the elevation map on which entire analysis is based.
An optional input can be:
· Depression map: Map layer of actual depressions in the landscape that are large enough to slow and store surface runoff from a storm event. Any non-zero values indicate depressions.
· Flow map: Amount of overland flow per cell. This map indicates the amount of overland flow units that each cell will contribute to the watershed basin model. Overland flow units represent the amount of overland flow each cell contributes to surface flow. If omitted, a value of one (1) is assumed.
· disturbed land map: Raster map input layer or value containing the percent of disturbed land (i.e., croplands, and construction sites) where the raster or input value of 17 equals 17%. If no map or value is given, r.watershed4.0 assumes no disturbed land. This input is used for the RUSLE calculations.
· blocking map: terrain that will block overland surface flow. Terrain that will block overland surface flow and restart the slope length for the RUSLE. Any non-zero values indicate blocking terrain.
· threshold value: The minimum size of an exterior watershed basin in cells, or overland flow units.
· max.slope.length map: Input value indicating the maximum length of overland surface flow in meters. If overland flow travels greater than the maximum length, the program assumes the maximum length (it assumes that landscape characteristics not discernible in the digital elevation model exist that maximize the slope length). This input is used for the RUSLE calculations and is a sensitive parameter.
Output maps are:
· accumulation map: number of cells that drain through each cell. The absolute value of each cell in this output map layer is the amount of overland flow that traverses the cell. This value will be the number of upland cells plus one if no overland flow map is given. If the overland flow map is given, the value will be in overland flow units. Negative numbers indicate that those cells possibly have surface runoff from outside of the current geographic region. Thus, any cells with negative values cannot have their surface runoff and sedimentation yields calculated accurately.
· drainage direction map: Provides the "aspect" for each cell. Multiplying positive values by 45 will give the direction in degrees that the surface runoff will travel from that cell. The value -1 indicates that the cell is a depression area (defined by the depression input map). Other negative values indicate that surface runoff is leaving the boundaries of the current geographic region. The absolute value of these negative cells indicates the direction of flow.
· basin map: Unique label for each watershed basin. Each basin will be given a unique positive even integer. Areas along edges may not be large enough to create an exterior watershed basin. 0 values indicate that the cell is not part of a complete watershed basin in the current geographic region.
· stream map: stream segments. Values correspond to the watershed basin values.
· half.basin map: each half-basin is given a unique value. Watershed basins are divided into left and right sides. The right-hand side cell of the watershed basin (looking upstream) are given even values corresponding to the watershed basin values. The left-hand side cells of the watershed basin are given odd values which are one less than the value of the watershed basin.
· visual map: useful for visual display of results. Surface runoff accumulation with the values modified to provide for easy display. All negative accumulation values are changed to zero. All positive values above the basin threshold are given the value of the basin threshold.
· length.slope map: slope length and steepness (LS) factor. Contains the LS factor for the Revised Universal Soil Loss Equation. Equations taken from Revised Universal Soil Loss Equation for Western Rangelands (see SEE ALSO section). Since the LS factor is a small number, it is multiplied by 100 for the GRASS output map.
· slope.steepness map: slope steepness (S) factor for RUSLE. Contains the revised S factor for the Universal Soil Loss Equation. Equations taken from article entitled Revised Slope Steepness Factor for the Universal Soil Loss Equation (see SEE ALSO section). Since the S factor is a small number (usually less than one), it is multiplied by 100 for the GRASS output map layer.
r.water.outlet - Subwatershed basin creation program.
r.water.outlet generates a watershed basin from a drainage direction map (from r.watershed) and a set of coordinates representing the outlet point of this particular watershed. In this way catchment areas can be calculated for each particular point in a DEM.
r.basins.fill - Generates a raster map layer showing watershed subbasins.
r.basins.fill generates a raster map layer depicting subbasins, based on input raster map layers for the coded stream network (where each channel segment has been "coded" with a unique category value) and for the ridges within a given watershed. The raster map layer depicting ridges should include the ridge which defines the perimeter of the watershed. The coded stream network can be generated as part of the r.watershed program, but the map layer of ridges will need to be created by hand, either through digitizing done in v.digit, or through the on-screen digitizing option accessible within d.display. The resulting output raster map layer will code the subbasins with category values matching those of the channel segments passing through them. A user-supplied number of passes through the data is made in an attempt to fill in these subbasins. If the resulting map layer from this program appears to have holes within a subbasin, the program should be rerun with a higher number of passes.
r.mask - Establishes or removes the current working mask.
The r.mask module allows the user to block out certain areas of a map from analysis, by "hiding" them from sight of other GRASS programs. This is done by establishing a mask. While a mask exists, most GRASS programs will operate only on data falling inside the masked area, and ignore any data falling outside of the mask. In watershed modelling the area outside the watershed can be masked.
r.mask.points -Examines and filters lists of points constituting lines to determine if they fall within current region and mask and optionally an additional raster map.
r.mask.points filters a list of points based on the current region and current mask. The point list consists of lines which have the following format
easting northing [text]
... ... ...
easting northing [text]
The eastings and northings define points in the coordinate space. Each line is examined to determine if the point falls within the current region, current mask (if any), and optionally an additional raster map that acts as a secondary mask. If the point falls outside the current region or falls in a grid cell that has value zero (either in the current mask, or the specified mask file), then the entire line is suppressed. Otherwise it is printed exactly as it is input. There may be arbitrary text following the coordinate pairs and this text is output as well.
12.3.1 Contour lines / DEM surfaces
r.contour - Produces a vector map of specified contours from raster DEM map layer.
The module r.contour produces a contour map of user-specified levels from a raster map layer. This program works two ways:
1. Contours are produced from a user-specified list of levels.
2. Contours are produced at some regular increment from user-specified minimum level to maximum level. If no minimum or maximum level is specified, minimum or maximum data value will be used.
r.surf.contour - Surface generation program from rasterized vector contours.
r.surf.contour creates a raster elevation map from a rasterized contour map (v.to.rast). Elevation values are determined using procedures similar to a manual methods. To determine the elevation of a point on a contour map, an individual might interpolate its value from those of the two nearest contour lines (uphill and downhill). r.surf.contour works in a similar way. Initially, a vector map of the contour lines is made with the elevation of each line as its label (see v.digit ). When the program v.to.rast is run on the vector map, continuous "lines" of rasters containing the contour line values will be the input for r.surf.contour. For each cell in the input map, either the cell is a contour line cell (which is given that value), or a flood fill is generated from that spot until the fill comes to two unique values. The flood fill is not allowed to cross over the rasterized contour lines, thus ensuring that an uphill and downhill contour value will be the two values chosen. r.surf.contour interpolates from the uphill and downhill values by the true distance.
r.surf.idw/r.surf.idw2 - Surface interpolation utility for raster map layers for missing values.
r.surf.idw(2) fills a grid cell (raster) matrix with interpolated values generated from a set of input layer data points. It uses a numerical approximation technique based on distance squared weighting of the values of nearest data points. The number of nearest data points used to determined the interpolated value of a cell can be specified by the user (default: 12 nearest data points).
r.surf.idw is used for lat./log. locations while r.surf.idw2 for projection locations in UTM etc.
s.surf.idw - Surface generation from sites data program (spot heights).
s.surf.idw fills a raster matrix with interpolated values generated from a set of irregularly spaced data points using numerical approximation (weighted averaging) techniques. The interpolated value of a cell is determined by values of nearby data points and the distance of the cell from those input points. In comparison with other methods, numerical approximation allows representation of more complex surfaces (particularly those with anomalous features), restricts the spatial influence of any errors, and generates the interpolated surface from the data points. It is the most appropriate method to apply to most spatial data.
12.3.2 Flowpath, depression lines, flow direction maps, slope, aspect maps
r.direct - Generates a flow direction map from a given elevation layer
The type of format can be chosen which the user wishes to create the flow direction map. The agnps format gives category values from 1-8, with 1 facing north and increasing values in the clockwise direction. The answers format gives category values from 0-360 degrees, with 0 (360) facing east and values increasing in the counter clockwise direction at 45 degree increments. The grass format gives the same category values as the r.slope.aspect program.
r.drain - Traces a flow through an elevation model on a raster map layer.
r.drain traces a flow through a least-cost path in an elevation model. The elevation surface (a raster map layer input) might be the cumulative cost map generated by the r.cost program. The output result (also a raster map layer) will show one or more least-cost paths between each user-provided location(s) and the low spot (low category values) in the input model.
r.fill.dir - Filters and generates a depressionless elevation map and a flow direction map from a given elevation layer
Parameters:
input = elevation map
elevation = corrected elevation map
direction = flow direction map
type = type of flow direction map to be created (AGNPS, ANSWERS, GRASS, see r.direct for explanation)
r.flow - construction of slope curves (flowlines), computation of raster maps of flowpath length and flowline densities from a raster digital elevation model
This program generates flowlines using combined raster-vector approach from input elevation and aspect raster maps. The output is a vector file of flowlines and/or a raster map of flowpath lengths and/or a raster map of flowline densities. Flowlines constructed by this program are represented in the vector format, given by coordinates of the intersections of the flowline with edges of the mesh which connects centers of grid cells. Aspect used for input must follow the same rules as aspect computed in other GRASS 4.2 programs (see r.slope.aspect , s.surf.tps ).
Flowlines are generated from each cell uphill by default, they can be generated downhill using the flag -d. Flowline stops when it reaches a point with lower or equal (for uphill case), higher or equal (for downhill case) elevation value, or a barrier (e.g.road). Barriers are optionally defined by non-zero values in an input raster map barierin.
Flowpath length flout for each grid cell is computed as a plan distance summing lengths of linear parts of the flowline between intersections. Flowline densities dsout represent a number of flowlines passing through the given cell. Flowline lengths and densities can be computed for both uphill and downhill flowlines.
r.slope.aspect - Generates raster map layers of slope and aspect from a raster map layer of true elevation values.
r.slope.aspect generates raster map layers of slope and aspect from a raster map layer of true elevation values. The user must specify the input elevation file name and at least one output file name to contain the slope or aspect data. The user can also specify the format for slope (degrees, percent; default=degrees), and zfactor: multiplicative factor to convert elevation units to meters; (default 1.0).
d.profile - Displays profiles of a user-specified raster map layer.
This command works only interactively. It clears the entire graphics screen and provides a graphical interaction allowing the selection of transects for which profiles are then generated.
First, you will be presented with a prompt asking you to choose a raster map layer to be profiled. Once you specify a valid raster map layer name, the map layer will be displayed in the left half of the graphics display, and the right half of the dispay will be divided into four display frames. There will also be two frames along the top of the display: a mouse-button menu frame on the left, and a status frame on the right.
r.profile - Outputs the raster map layer values lying on user-defined line(s).
This module outputs, in ASCII, the values assigned to those cells in a raster map layer that lie along one or more lines ("profiles"). The lines are described by their starting and ending coordinates. The profiles may be single-cell wide lines, or multiple-cell wide lines. The output, for each profile, may
be the category values assigned to each of the cells, or a single aggregate value (e.g., average or median value).
r.transect - Outputs raster map layer values lying along user defined transect line(s).
This module outputs, in ASCII, the values in a raster map which lie along one or more user-defined transect lines. The transects are described by their starting coordinates, azimuth, and distance. The transects may be single-cell wide lines, or multiple-cell wide lines. The output, for each transect, may be the values at each of the cells, or a single aggregate value (e.g., average or median value).
r.volume - Calculates the volume of data "clumps", and (optionally) produces a GRASS site_lists file containing the calculated centroids of these clumps.
Parameter:
data=elevation (The cell resolution (area) of the data map will also be used.)
[clump=clumplayer] Name of an existing raster map layer that defines the boundaries of each clump. Preferably, this map should be the output of r.clump. If the user has imposed a mask, the program uses this mask as the clump map layer if no other clump layer is specified by the user.
[site_list=sitesfile] The name to be assigned to a new GRASS site_list file, in which clump centroids can be stored.
For ease of example, it is assumed that each clump is a field, that cell category values in the elevation map layer represent actual elevation values in meters, and that the data base is a UTM data base (in meters). This means that a field (clump A) contains an amount of cells; the average elevation in field A is a second value. The sum of all of the elevation values assigned to cells within field A the third value. The volume (x*y*z) of space in field A is equal to the volume.
Very interesting applications can be done with r.volume. For instance, one can calculate: the volume of rock in a potential quarry; cut/fill volumes for roads; and, water volumes in potential reservoirs. Data layers of other measures of real values can also be used.
12.4 Rainfall data preparation
s.surf.idw - Generation of Thiessen polygons from sites data (rainfall point data) into raster format.
s.surf.idw fills a raster matrix with interpolated values generated from a set of irregularly spaced data points using numerical approximation (weighted averaging) techniques. The interpolated value of a cell is determined by values of nearby data points and the distance of the cell from those input points.
If the number of interpolation points is set to 1 the module calculates Thiessen polygons and stores them in a new raster layer.
s.voronoi - Generation of Thiessen polygons from sites data (rainfall point data) into vector format.
The module s.voronoi calculates vector Thiessen polygons from given rainfall point data.
12.5 Erosion and transportation models
r.cn - Generates a curve number map layer
r.cn is a command-line interface for generating an SCS CN map. Input parameters are
sg=map hydrologic soil group map name
lu=map landuse map name
pr=map practice or management map name
hc=map hydrologic condition map name
cn=map output CN map name
amc=number equivalent AMC condition number for the CN output
r.weighted.cn - Generates a weighted SCS curve number map layer from a CN-map calculated with r.cn module.
r.agnps - Menu-driven interface from GRASS to AGNPS 5.0
The WATERSHEDSS GRASS-AGNPS modeling tool was developed to assess nonpoint source pollution origination and movement in a watershed. Users with the appropriate GRASS map layers can simulate nutrient, sediment, and pesticide loads associated with various land management/use scenarios by running the input file generator and AGNPS model contained in the tool. Current land management/use within the watershed can be evaluated to identify source areas that contribute relatively high levels of pollutants to various points in the watershed. Other land management/use scenarios such as changing or moving cropping systems to different areas on the landscape may be assessed by modifying GRASS map layers and running the tool. Simulated pollutant yields from various scenarios may then be compared to provide estimates of pollutant reductions associated with changes in land management / use. There is an extra manual available for this module.
r.answers - Menu-driven interface from GRASS to ANSWERS
r.answers integrates ANSWERS with GRASS. ANSWERS (Areal Nonpoint Source Watershed Environmental Response Simulation) is an event oriented, distributed parameter model that was developed to simulate the behavior of watersheds having agriculture as their primary land use. Its primary applications are watershed planning for erosion and sediment control on complex watersheds, and water quality analysis associated with sediment associated chemicals. There is an extra manual available for this module.
r.runoff - Generates an SCS curve number runoff map layer. It is a command-line interface for generating the runoff map by the SCS method.
Parameters:
rf = rainfall map name in mm
cn = CN map name
ro = output runoff map name in *100 mm
r.water.fea - Finite element analysis program for hydrologic simulations.
r.water.fea is an interactive program that allows the user to simulate storm water runoff analysis using the finite element numerical technique. Infiltration is calculated using the Green and Ampt formulation. r.water.fea computes and draws hydrographs for every basin as well as at stream junctions in an analysis area. It also draws animation maps at the basin level. There is an extra manual available for this module.
12.7 Calculations on raster layers
r.mapcalc - Raster map layer data calculator.
The powerful module r.mapcalc performs arithmetic on raster map layers. New raster map layers can be created which are arithmetic expressions involving existing raster map layers, integer or floating point constants, and functions.
r.patch - Creates a composite raster map layer by using known category values from one (or more) map layer(s) to fill in areas of "no data" in another map layer.
The GRASS module r.patch allows the user to assign known data values from other raster map layers to the "no data" areas (those assigned category value 0) in another raster map layer. This program is useful for making a composite raster map layer from two or more adjacent map layers, for filling in "holes" in a raster map layer's data (e.g., in digital elevation data), or for updating an older map layer with more recent data.
r.rescale - Rescales the range of category values in a raster map layer.
The r.rescale program rescales the range of category values appearing in a raster map layer. A new raster map layer, and an appropriate category file and color table based upon the original raster map layer, are generated with category labels that reflect the original category values that produced each category. This command is useful for producing representations with a reduced number of categories from a raster map layer with a large range of category values (e.g., elevation). Rescaled map layers are appropriate for use in such GRASS programs as r.stats, r.report, and r.coin.
r.reclass - Creates a new map layer whose category values are based upon the user's reclassification of categories in an existing raster map layer.
r.reclass creates an output map layer based on an input raster map layer. The output map layer will be a reclassification of the input map layer based on reclass rules input to r.reclass, and can be treated in much the same way that raster files are treated. A title for the output map layer may be (optionally) specified by the user.
r.reclass.scs - Create a new raster map layer based on an existing raster map.
r.reclass.scs is an interface to the GRASS r.reclass program. The program will reclassify the category values in a raster map layer based on reclass instructions entered by the user. The user can enter map reclassification rules to r.reclass.scs either from standard input or from a file. The program then issues r.reclass commands to produce the new reclassified raster map layer.
12.8 Automated boundary and line digitizing
r.line - Creates a new binary GRASS vector (v.digit) file by extracting linear features from a thinned raster file.
r.line scans the named raster map layer (input=name) and extracts thinned linear vector features into the named vector file (output=name). It works only with thinned raster layers (use r.thin). The user must run v.support on the resultant vector files to build the dig_plus information. r.thin and r.line may create excessive nodes at every junction, and may create small spurs or "dangling lines" during the thinning and vectorization process. These nodes and spurs may be removed using v.trim.
r.poly - Extracts area edges from a raster map layer and converts data to GRASS vector format.
r.poly scans the named input raster map layer, extracts area edge features from it, converts data to GRASS vector format, and smoothes vectors.
r.poly first traces the perimeter of each unique area in the raster map layer and creates vector data to represent it. The cell category values for the raster map layer will be used to create attribute information for the resultant vector area edge data.
A true vector tracing of the area edges might appear blocky, since the vectors outline the edges of raster data that are stored in rectangular cells. To produce a better-looking vector map, r.poly smoothes the corners of the vector data as they are being extracted. At each change in direction (i.e., each corner), the two midpoints of the corner cell (half the cell's height and width) are taken, and the line segment connecting them is used to outline this corner in the resultant vector file. (The cell's cornermost node is ignored.) Because vectors are smoothed by this program, the resulting vector map will not be "true" to the raster map from which it was created. The user should check the resolution of the geographic region (and the original data) to estimate the possible error introduced by smoothing. The user must run v.support on the resultant vector files to build the dig_plus information.
r.thin - Thins non-zero cells that denote linear features in a raster map layer.
r.thin will thin only the non-zero cells of the named input raster map layer within the current geographic region settings. The cell width of the thinned output raster map layer will be equal to the cell resolution of the currently set geographic region. All of the thinned linear features will have the width of a single cell.
r.thin will create a new output raster data file containing the thinned linear features. r.thin assumes that linear features are encoded with positive values on a background of 0's in the input raster data file.
Using satellite images and aerial photos, a large number of environmental questions can be answered. Especially in areas, where no thematic maps are available, these images are very helpful. Digital images consist of pixels, in GIS these pixels are the same as raster cells. A pixel is the smallest information unit in an image.
After the recification of a raw image into a coordinate system the different bands (channels) of a satellite image can be overlayed (so-called color composites). The former greyscale images result in
a new color image. There the landcover like the vegetation coverage, waterbodies etc. or the landuse
is emphasized through the colors. Usually false colors are generated. Other calculations may be band ratios, band differences etc. Using mathematical algebra a variety of information can be found out.
Improvements of the original images can be performed by geometrical and radiometrical filtering of the image. GRASS offers all these features, especially in image enhancement there are no limitations to define own filters.
All descriptions given here are valid for satellite images as well as aerial photos.
1.1 Definition of the xy-location for unrectified images
The raw satellite images and aerial photos have to be imported into a xy-location first. Therefore create this location following the instructions of the first handout. Choose the xy-system after giving a new location name and specify in the "region definition screen" the xy-coordinates.
Example for a image with size: 512 * 1200 (x=512, y=1200):
1.2 Import of satellite images
After creating the location and the mapset you can start to import the data. Two possibilities are there:
· import in standard image format (GIF, TIFF, PPM, SUN Raster format)
· import in BIL/BSQ tape format
1.2.1 Import in standard image formats
The import in this format is rather easy, follow the description given in "raster data management". After using the import module (
r.in.gif, r.in.tiff, r.in.ppm) the module r.support has to be used on all bands to set the coordinates properly ("edit header": y) and to update the histograms.
1.2.2 Import in BIL/BSQ tape format
The raw data are mostly delivered on tapes or nowadays on CDROM. Two main storing formats are used: BIL (band interleave format) and BSQ (band sequential format). In a unix machine (therefore also in LinuX) these tape- and CDROM-drives are implemented as "devices". In the directory
/dev/
must be a device like (examples here!)
/dev/cdrom for CDROM
/dev/rmt0 for the first tape drive (for a density of 1600)
/dev/fd0 for the first floppy drive
etc. This is system specific, therefore ask you system operator.
To check the medium, if you are not shure about the parameters, you can use:
$ m.examine.tape
enter tape device name: /dev/rmt0
Enter size of buffer to use when reading the tape [32767]
Please mount and load tape, then hit <RETURN>:
Results to a file? (y/n) n
The best way is to define the format youself in the import module, relying on the known tape-parameters. So you use:
$ i.tape.other
enter tape device name: /dev/rmt0
Enter size of buffer to use when reading the tape [32767]
Into the following screen you can enter a free description:
Then the tape-specific parameters follow (example for BIL 512 * 512, 4 Band scene):
After leaving this screen with <ESC> you mark a "x" for each channel of interest (mostly all). This is followed by the option to extract only a particular area out of all channels. To get the entire scene give
1 for minimum value and the record length for the maximum value:
Example:
start row: 1______
end row: 512____
start col: 1______
end col: 512____
after specifying the name of the image scene the import process begins. Use (as usual)
r.support after importing the images.
2. Rectification of satellite images and aerial photos
To rectify the satellite images or aerial photos into a projection like UTM you first have to create this as a new target location. Satellite images can be rectified into this new location with an affine or polynomial transformation, aerial photos with an ortho rectification onto a DEM.
So you leave GRASS after the import of the images into the xy-location and start GRASS again either with a new name for the location and mapset or with an already existing location/mapset. In the first case define your coordinates, projection and ellipsoid and the resolution. The resolution should be set to the minimum resolution you want to use in this location (depends on your input data like DEM, scanned maps etc.). This resolution is the default resolution. After defining the database you get into the system. The same certainly takes place if the location is already defined. Now you have to prepare the location for the satellite recification. So you set the coordinates to the coordinates the satellite image or the aerial photo shall get later. Also the resolution has to be set for the specific image. It may differ from the default resolution, but can only be equal or higher than this. You set the resolution and coordinates with
$ g.region
Then leave GRASS (this new coordinates and will be stored) and start GRASS again calling the xy-location.
2.1 Collect the image bands in a group
The first step is now to create a "group". Here all images will be specified, which shall be transformed into the projection-location.
The module:
$ i.group
collect the bands of interest (mark them with an "x").
A subgroup is not necessary here (that is only necessary for the classification module later).
2.2 Specifying the target location
GRASS has to know the target location into the images shall be transformed. With the module:
$ i.target
you can specify the target location and the mapset. Please don't forget that you have to set the target coordinates and the image resolution, as described in the beginning of the chapter. Otherwise the parameters won't fit and the result will be bad.
2.3 Setting the ground control points (GCPs)
Now the procedure of setting the GCPs follows. To set the GCPs there are three possibilities:
· Raster to raster setting of GCPs:
Display the unrectified image and a raster image from the target location to set the GCPs. This is only possible, if you transform the image into a existing target location with maps. Then you can use the mouse to set the GCP in the unrectified image and the corresponding point in the target location.
The module's name is:
· Raster to vector setting of GCPs:
Display the unrectified image and a vector image from the target location to set the GCPs. This is only possible, if you transform the image into a existing target location with maps. Then you can use the mouse to set the GCP in the unrectified image and the corresponding point in the target location.
The module's name is:
$ i.vpoints· Raster to given coordinates setting of GCPs:
Display the unrectified image, set the GCPs with the mouse and type in the corresponding coordinates manually into the terminal. First you click the GCP in the image, then type in the easting and the northing with blank space in between.
The module's name is again:
$ i.pointsThese modules are self explaining, the menubar allows the choice of the image, zooming and the setting of the points. To make the finding of GCPs easier you should make a copy of one image with
$ g.copy
and perform a contrast stretch using
$ i.grey.scale
on that copy. This image can be displayed in i.points or i.vpoints. The GCPs are valid for all images in the "group".
2.4 Verifying the quality of GCPs
To check if the GCPs are set properly, use the
Analysis-function
from the menubar. Then the RMS-error (root mean square error) is calculated. With red color the bad GCPs are marked, you can disable them. After getting a good result you can leave this module. The points will be saved. The summated RMS-error (last line in the table) shall be less than the ground resolution.
The minimum number of GCPs depends on the transformation algorithm you want to use. You need for
· Affine transformation: 4 GCPs in the corners
· Polynomial transformation: It depends on the order of the polynom (p)
(p + 1) * (p + 2)
number of GCPs: -----------------------
2
For a transformation 3rd order 10 GCPs are necessary. But you should give more GCPs for polynomial transformation to calculate the RMS-error better.
The transformation which has to be used depends on the image source. For satellite images normally the polynomial transformation has to be used, because there is geometrical distortion in the image itself. For images which only have to be rotated and stretched you use the affine transformation.
The module for the affine transformation is:
$ i.rectify
Transform the images into the "current region" because you have set it in your target location properly. The module works in the background and sends an email after finishing the transformation. You can leave GRASS in the meanwhile if you want and logout also.
2.6.2 Polynomial transformation
The module for the polynomial transformation is:
$ i.rectify2
First you specify the order of the polynom (usually 3), then the "current region" as target. The module works in the background and sends an email after finishing the transformation. You can leave GRASS in the meanwhile if you want and logout also.
2.6.3 Ortho photo rectification
For aerial photos you need DEM in the target location and the coordinates of the nadir point (must not be too exact). Also the flight parameters are required (altitude, camera parameters etc.). Then you enter the xy-location and start the module:
$ i.ortho.photo
It is menu driven – you have to follow all steps from the top. For detailed information read the man-page of i.ortho.photo.
Using the images
After the finish of the transformation you should start GRASS with the target location and check, if the transformation was done properly. In this case you can delete the xy-location (it is a directory in the GRASS database-directory):
$ cd grass_data
$ rm -rf <locationname>
Now you can perform image enhancements, calculate color-composites etc.
After the transformation of the satellite image or aerial photo the images finally "arrive" in the location with projection (UTM etc.). The bands are still in raw format, only rectified. This was the geometric correction of the images. The enhancement is the radiometric correction - the frequencies, colors etc. are modified. Enhancements can be done in several ways: contrast stretching, filtering, edge detection, fourier transformation and so on.
To possibilities have to be distinguished: You can either
· modify the colortable or
· modify the cell values
Both possibilities are discussed now.
To perform a contrast stretch you can choose between changing only the colortable or affect the cell values.
Stretch the color table
To stretch only the colortable but leave the image untouched you can use the module:
$ i.grey.scale
It has to be used interactively and assigns a new colotable to each specified band.
Stretch the cell values
The real stretch of the cell values, which has an effect on further calculations can be done with
$ r.rescale or
$ r.mapcalc
For a linear stretch you can use the first module which is described above in the "raster management" chapter. The more complex non-linear stretch needs the second module. To get the minimum and maximum values of the band you use:
$ r.stats -c <bandname>
The first value (first column) indicates the minimum, the last value (first column) the maximum cell value in this image. Then you can do your calculations in
r.mapcalc.
3.2 Principal and canonical component analysis
The PCA allows to "concentrate" the information stored in all image bands in the first bands. The information content is decreasing to the higher band numbers. Also the used space is less. The mo-dule rotates the internal x and y axis of the images and stores the information in new raster files.
The module is:
$ i.pca
First all input bands have to be given, then one additional return, after that the prefix for the output bands (they get numbers by the module). Sometimes it is better to work on these principal component transformed images.
Using spectral signatures (these signatures can be created from training areas with
i.class, see later on) you can also perform a canonical component analysis. The module is used in the same manner:$ i.cca
3.3 Fourier and inverse Fourier transformation
A very interesting tool for radiometric analysis and corrections is the Fourier-transformation-module. It converts an image into its spatial frequency domains (the complex function results: real and imaginary frequency components) so that its spectral distribution is displayable. After this conversion specific frequencies can be masked out. When the image is backtransformed into geographic coordinates these frequencies will be missing in the backtransformed image. In that way you can eliminate distortions (e.g. line distortion in satellite images) or filter certain frequencies:
· low frequencies
· high frequencies
· create band filters
The theory of fourier transformation is well explained in JENSEN 1996.
The window size must be square and a power of 2 in size (e.g., 64, 128, 256, ...). This is due to the mathematical properties of the algorithms used. GRASS take care of this itself.
Fourier transformation
First you transform the image into the real and imagery frequency components using
$ i.fft
You specify the image name and two new names for these new images. Then you can display the real part.
The structure in this FFT-image is like this:
Interpretation of frequency transformed images can be quite complicated. The output image (usually only the real image is used for visual analysis, but both parts are required for the backtransformation) is the two-dimensional frequency spectrum of the input image. The result is fairly symmetrical. Frequencies are along both axis (X and Y). The DC point (Direct Current, frequency is 0,0) is located at (S/2+1, S/2+1) where S is the image size. Example: A 512 x 512 pixel image would have the DC point at the coordinates x=257, y=257. Starting with the lowest frequencies the points away from the DC point indicate higher frequencies. Also colors are displayed: The brightness of a point indicates the number of times that frequency exists in the original image.
Creating filters
The question is now to filter this image. Specific frequencies can be filtered out using masks: The basis for the filter technique is to eliminate or reduce some frequencies while keeping others. The filtering can be performed by creating a mask consisting of "0", where frequencies should be eliminated, and "1" where they should be kept. If this mask is multiplied with the fourier transformed bands, the results are two new image (still real and imaginary part) with undesirable frequencies eliminated. When the inverse transformation is applied on these corrected images, the result is the original image with certain frequencies removed. For example, the result of keeping only high frequencies would be the enhancement of edges. This multiplying of the mask with the image is done internally by GRASS (module
r.mask).
Filters are usually symmetric around the DC point which is at (S/2+1, S/2+1), where S is the image size. To create this mask use the module
$ r.digit
There is a feature "Circle". With the mouse one circle can be drawn, it shall get the category value 1 and the label "inner circle". This circle will be saved as raster image while leaving the module.
Now the mask has to be overlayed:
$ r.mask
Specify the name of the "circle"-rasterfile. The choice is now to specify the value one (active area) and the value 0 (inactive area). Example: In a low pass filter the "inner circle" becomes 1, the outer area 0.
Some common filters are ("filled" means: specify value 1 in
r.mask):
+----------------+ +----------------+
| | |XXXXXXXXXXXXXXXX|
| | |XXXXXXXXXXXXXXXX|
| XXXXX | |XXXXXX XXXXX|
| XXXXXXX | |XXXXX XXXX|
| XXXXXXX | |XXXXX XXXX|
| XXXXXXX | |XXXXX XXXX|
| XXXXX | |XXXXXX XXXXX|
| | |XXXXXXXXXXXXXXXX|
| | |XXXXXXXXXXXXXXXX|
+----------------+ +----------------+
Low Pass filter High Pass Filter
(Filled Circle) (Filled outside Circle)
Inverse Fourier transformation (backtransformation)
After setting the mask, the image has to be backtransformed (inverse Fourier transformation):
$ i.ifft
Here again the both images (real and imagery part) have to be specified and a new name for the filtered resulting image.
Very important is to remove the mask after the inverse Fourier transformation using
$ r.mask
again, otherwise the mask is also valid for the display of geographic raster maps (the "normal" raster maps). Now the filtered image can be displayed.
You can always use this filter also reversed, you just have to reverse the category values 1 and 0 in
r.mask.
More complex filters
If you want to have two circles or you want to filter out non circular parts (e.g. to eliminate distortion) the "Circle" of
r.digit cannot be used. The outer circle would overlay the inner circle. So you will make use of the feature "Area". The digitizing has to be done like this:Digitize the inner circle with a lot of nodes to approximate it (so the circle is now consisting of lines), after reaching the starting point of the circle digitize further on to the radius of the outer circle, digitize the outer circle (so don't stop digitizing in between) and stop digitizing at the starting point of the outer circle. The area between the circles will be "filled", if you specify the the category value 1.
Then you can leave the
r.mask-module.
For controlling you should display this raster-"filter"-file.
The band pass and band cut filters look like this:
+----------------+ +----------------+
| | |XXXXXXXXXXXXXXXX|
| XXXX | |XXXXXX XXXXXX|
| XXXXXX | |XXXXX XXXXX|
| XXX XXX | |XXXX XX XXXX|
| XX XX | |XXXX XXXX XXXX|
| XXX XXX | |XXXX XX XXXX|
| XXXXXX | |XXXXX XXXXX|
| XXXX | |XXXXXX XXXXXX|
| | |XXXXXXXXXXXXXXXX|
+----------------+ +----------------+
Band Pass Filter Cut Pass Filter
(Donut) (Filled Outside Donut)
Distorted images
If you have line distortion in a satellite image, the FFT-transformed image will have a shape like the left example.
+----------------+ +----------------+
| | | | | | |
| | | | | | |
| |XXX| | | X X |
| XXXXXXX | | XXXXXXX |
| XXXXXXX | | XXXXXXX |
| XXXXXXX | | XXXXXXX |
| |XXX| | | X X |
| | | | | | |
| | | | | | |
+----------------+ +----------------+
FFT of distored FFT of distored image
image after masking out the
distortion
Distortion from stationary periodic noise becomes a singe bright point in the Fourier transform. This can be easily remove in the transformed image but only with difficulties in the original image.
Stripes in the FFT will occur when periodic noise of various frequencies is there (example above).
With area-masking you can eliminate the distortion (right example) and transform it back with
$ i.ifft
GRASS offers a tool for the self-defining of matrix filters. These filters are a moving matrix window, which moves over the raster image and does some calculation on all raster cells within this window. This can be used for
· low and high pass filtering
· averaging
· edge detection
· resampling (changing the resolution)
Two ways of defining these filter are there: Using
r.mapcalc and using r.mfilter. The first way is more complex because the neighbour cells from the center cell have to be defined with relative coordinates (e.g. the cell upper left is [-1,1], the center cell [0,0]). More information about this you find in the r.mapcalc-analysis tutorial.Easier is to use the module
$ r.mfilter
You can define you filter in an ASCII file which has to be stored in the working directory.
The first line is a title, the second the size of the matrix, then the matrix itself follows, then a divisor and finally the type of filter (sequential or parallel).
Example (the keywords have to be specified):
TITLE Edge enhancement
MATRIX 3
-1 0 -1
0 -8 0
-1 0 -1
DIVISOR 9
TYPE P
In this example the center cell is weighted with -8, the surrounding cells with different values. The entire matrix is divided through 9.
In that manner all types of filters can be defined, the only limitation is that the numbers have to be whole numbers. In
r.mapcalc this limitation is not there.
A helpful feature is to overlay images to get a resulting false colors image. You assign here one band to each basic color (red, green, blue). With high band images (LANDSAT TM) there is a big variety of compositions.
To calculate these composites you first have to collect the images in a group:
$ i.group
A subgroup is not necessary. The composites itself can be defined in
$ i.composite
Specifiy the letter r,g and b for the desired bands. The number of color levels should be 7or 8 (the resulting number of colors is (3 power of "level"). After giving a name for the composite the calculation is performed.
Then you can display the color composite.
Here a list of band and color composite descriptions follows (after Kevin Seel, seel@fsa.ca):
LANDSAT-TM 5 Spectral Bands (30m resolution):
1 blue (0.45-0.53 um)
Recommended for use in combination with other bands because of low contrast
and high sensitivity to haze.
2 green (0.52-0.60 um)
Recommended for use in combination with other bands because of low contrast
and high sensitivity to haze.
3 red (0.63-0.69 um)
Best for showing roads and bare soils. This band heightens the contrast between
vegetated and non-vegetated areas.
4 near-infrared (0.76-0.90 um)
Useful for biomass estimation. It also separates water bodies from vegetation.
Not as effective as band 3 for identifying roads.
5 shortwave infrared (1.55-1.75 um)
Best single overall band. It shows roads, openings, bare soils, water
good contrast between different types of vegetation. It also has excellent haze
penetration.
6 thermal infrared (10.5-12.5 um)
This band responds to thermal radiation emitted by the target. Thermal IR is
closely related to soil moisture and the height and temperature of the vegetation.
Data are acquired at night to obtain maximum information. (note 80m res for
this band only)
7 shortwave infrared (2.08-2.35 um)
This band has greater haze penetration than band 5. It is less useful than band 5 for
interpreting vegetation cover, but has strong potential for soil analysis.
Colour Composits in BGR order:
Example: 234 in BGR order means: B=2, G=3, R=4
234 Sensitive to green vegetation (portrayed as red), coniferous as distinctly darker
red than deciduous forests. Roads and water bodies are clear.
345 contains one band from each of the main reflective units (vis, nir, shortwave
infra). Green veg is green and the shortwave band shows stress and mortality.
Roads are less evident as band 3 is blue.
347 similar to 345 but depicts burned areas better.
243 green veg is green but coniferous forests arenUt as clear as the 234 combo
354 appears more like a colour infrared photo
374 similar to 354
123 true colour, however because of high correlation of the 3 bands in visible this
combo contains not much more info than contained in one band alone.
457 shows soil texture classes (clay, loam, sandy) by CCRS researchers.
247 one of the best for info pertaining to forestry. good for operation scale mapping
of recent harvest areas and road construction
Band ratioing is a another way to get specific information out of a satellite scene. The NDVI (normalized difference vegetation index) is calculated like that. The module
r.mapcalc can be used for calculating ratios. According to the limitation of whole numbers in raster images the formula should be multiplied with 10 or 100.
Example NDVI:
tm4 - tm3
ndvi = --------------
tm4 + tm3
mapcalc> ndvi = 10 * (tm4 - tm3) / (tm4 + tm3)
For this calculation you have to specify the raster layer names for tm4 and tm3 or use directly these names.
6. Introduction to satellite image classification
Satellite image scenes consist of several bands. These bands store the information about the spectral reflectance of the earth surface. In each band the pixel values will be different, depending on the measured frequency (which compares to the bands). Each band has a range of 0 to 255 greyscales which represent the spectral distribution within this band.
The idea of classification of satellite images is to categorize the contents of a satellite image scene into themes. Several bands of multispectral data are taken and analysed statistically. From the results one new image layer is calculated, containing the themes (or so-called classes) which are consisting of similar reflection of the ground surface. To these classes the landuse, waterbodies etc. can be manually assigned by connecting these patterns to ground information. Each pixel will be assigned to a class, a pixel is the same as a raster cell.
Using statistical methods on multispectral data (several bands) two ways of pattern recognition - the derivation of classes - can be performed:
a) spectral pattern recognition: Here the spectral distribution in the bands is analysed.
b) spatial pattern recognition: This recognition takes care of the geometric shapes and sizes of neighbourhood pixels.
GRASS offers both types of classification. Within these two types several ways of classifying an image are possible, depending on accuracy and time the user wants to spend for the calculation of the results:
a) The spectral pattern recognition:
· unsupervised classification: The image processing software performs the statistical analysis on the set of bands and derives the clusters itself. The algorithm takes care of given initial parameters (number of classes you finally want to have, some other statistical parameter) and calculates the statistical "clusterclouds". GRASS uses the "minimum distance to means" algorithm for deriving the clusters. In the second step the "maximum likelyhood" classifier is used to assign the pixels to classes: From each cluster a class is derived and all pixels are assigned. The classifier uses the cluster means and covariance matrices from the cluster-signature file.
· supervised classification: The supervised classification needs training areas defined by the user from which the spectral signatures are derived. From one area the first algorithm searches in the entire image for similar spectral reflectance and defines a cluster. Then the next area will follow. After the definition of all clusters the second step follows: The assignment of the pixels to the classes derived from the cluster with the pixel oriented "maximum likelyhood" classifier.
· semi supervised classification: In this classification the user will define some training areas which are well known (waterbodies, urban area, non-mixed forest etc.). These signatures will be put into the unsupervised classification as "seed signatures". Considering this the other clusters will be calculated. Then the "maximum likelyhood" classifier will be used.
The spectral pattern recognition can be either done before or after an image rectification because this classification as well as the rectification is pixel oriented.
b) spatial pattern recognition:
· supervised classification: The idea of this supervised classification is the same as in spectral pattern recognition. The user defines a training area map and cluster will be derived from this map. Then the "sequential maximum a posteriori (SMAP) estimation" algorithm assigns the pixels texture-oriented (contextual oriented) to the classes derived from the clusters.
Externally there are some other classifiers available, which not yet belong to the GRASS distribution. For example a "neural network" classifier is available in Internet. For more details please refer the "GRASS tutorial: image processing" and image processing books (see references).
The boundaries of the classified images can be automatically digitized into a vector layer (see chapter VI 12.2.7 "raster data management").
6.1 Spectral pattern recognition
The first step is to collect all images into an image group:
$ i.group
You define a group name and mark the images which shall be used for the classification with an "x". Then you have to define a "subgroup" (menue: 5.), give a name to the subgroup and again mark the images. After that you can leave this module.
6.1.1 Unsupervised classification
The unsupervised classification will be started with the "clustering" of the spectral information. The GRASS module is
$ i.cluster
The results of this clustering will be used in the "maximum likelyhood" classification. First you specify the name of group and subgroup containing the images.
Then the module asks for the "result signature": This is the file where all signatures found by the clustering algorithm will be stored. The "seed signatures" have to be empty, press <return> here. Now you should specify a report file. "Run in background" is not necessary.
The cluster-algorithm screen appears now:
Here the following information are required (items taken from GRASS man-page i.cluster):
Number of initial classes: The number of clusters that will initially be identified in the clustering process before the iterations begin. Specify for example 20 here. The following parameters can be untouched for a first trial.
Minimum class size: This is the minimum number of pixels that will be used to define a cluster, and is therefore the minimum number of pixels for which means and covariance matrices will be calculated. Default: 17
Class separation: This is the minimum separation below which clusters will be merged in the iteration process. If the separation is relatively large (1.5), clusters with similar spectral distributions will be merged. If the separation is small (0.5), clusters are more similar spectrally than the resulting clusters of a larger separation will be remain distinct. Smaller separations result in a fewer cluster mergers, while larger separations result in more mergers.
The optimum minimum class separation is an image is an image-specific number that depends on the image data being classified and the number of final clusters that are acceptable. Its determination requires experimentation. Commonly used minimum class separations range from 0.5 to 1.5. The separability matrix that is output via mail at the completion of the program presents the result of the minimum class separation.
Note that as the minimum class (or cluster) separation is increased, the maximum number of iterations should also be increased to achieve this separation with a high percentage of convergence (see percent convergence). This is because of the larger number of mergers that occur when separa-tion is larger. As more clusters are merged, the percentage decreases, requiring an increased number of stabilization iterations to achive a high percent convergence. Default: 0.0
Percent convergence: A high percent convergence is the point at which cluster means become stable during the iteration process. The default value is 98.0 percent. When clusters are being created, their means constantly change as pixels are assigned to them and the means are recalculated to include the new pixel. After all clusters have been created, i.cluster begins iterations that change cluster means by maximizing the distances between them. As these means shift, a higher and higher convergence is approached. Because means will never become totally static, a percent convergence and a maximum number of iterations are supplied to stop the iterative process. The percent convergence should be reached before the maximum number of iterations. If the maximum number of iterations is reached, it is probable that the desired percent convergence was not reached. In this event you may want to increase the number of iterations and run i.cluster again (see maximum number of iterations below). The number of iterations is available in mail and in the report file (see reportfile). Default: 98.0
Maximum number of iterations: This parameter determines the maximum number of iterations which is greater than the number of iterations predicted to achieve the optimum percent convergence. The default value is 30. If the number of iterations reaches the maximum designated by the user; the user may want to rerun i.cluster with a higher number of iterations (see reportfile). Default: 30
Sampling interval for rows and columns: These numbers are the default rows and columns skip, which are based on the size of the data set. They are computed to sample 10,000 pixels throughout each image band at 100 pixels per sampled row, and 100 pixels per sampled column. If the user changes the default intervals, and the sample size is too large for the available computer space, or too small for accurate analysis, a warning will be delivered by mail. The reason for sampling the data set is to save computer time and memory during the sampling process, while obtaining an accurate representation of the statistical range of data.
After leaving this screen the clustering will be done, each cluster is defined as one class.
Now the assignment of the pixels (raster cells) to the classes begins with "maximum likelyhood" classifier. Start the module:
$ i.maxlik
The module uses the cluster means and the covariance matrices from
i.cluster and determines to which cluster each pixel has the highest probability of belonging. This cluster becomes a class and the assigned pixel gets the class value (category of raster cell).First the group and subgroup have to be specified. Then the signature file follows: Enter here the filename you specified in
i.cluster as "result signature". After giving a name for the new raster layer for the classified image (the result) you can give a name for the "reject threshold map". That is a map which shows at which confidence level each pixel is likely to belong to the assigned class. In combination with r.mask you can mask out pixels with low confidence level (less probability of correct assignment). Two images are the result of this classification: The classified image and the reject threshold image.
6.1.2 Supervised classification
When the user specifies representative training areas the clustering rules are determined by this decisions. This training area is valid for all bands of a satellite image scene, in each band each particular raster cell (pixel) will have a different value. In that way the user defines the spectral signature him-/herself. After that the "maximum likelyhood" algorithm is used to assign the pixels to the derived classes.
To specify the training areas two different ways are possible:
1. Digitize the areas with a digitizer from a map (toposheet etc.), feed these areas into the clustering module and the resulting signatures into the classification algorithm.
2. Digitize the training areas directly in a image band or FCC using a GRASS module. The signatures are calculated directly, the histograms of all involved bands are displayed with marks for standard deviation of the distribution.
1. In the first case the user has to digitize the training areas using
$ v.digit in combination with
$ v.to.rast
or
$ r.digit
The resulting areas (don't forget to label these areas with values) have to be in raster format. This training map is the input for the module:
$ i.gensig
This module creates the signature file for
$ i.maxlik
Please refer chapter VII 7.1 for the use of
i.maxlik. The result is a classified image raster layer and a reject threshold map containing the probability of assignment of each pixel to its class.
2. The second way uses the module
$ i.class
after starting a GRASS monitor with
d.mon. After specifying group and subgroup the name for the "resulting signature file" has to be given (it is the input for i.maxlik). Then a "seed signature file" can optionally be given: It can be the "result signature file" of a training session before, if you stopped work and want to proceed now. In this case you would specify a new name for the "result signature file" for the actual session and give the "result signature file" from the previously session here as "seed signature file". In that way you can distribute your work on several sessions. If you start fresh, just press <return>.Then the user will be asked for a raster image to be displayed for defining the training areas. This might be a contrast stretched band or a FCC created with i.composite (see above in chapter VII 4).
Making use of the feature that the sessions can be interrupted, also different images can be used for finding the training areas (band ratios, NDVI etc.).
The module i.class is menu driven in the GRASS monitor. There is a ZOOM-option which is very useful. You start digitizing with the menue Define Region. There you click Draw Region and begin to digitize a polygon (e.g. a waterbody). After finishing the area you close it with Complete Region and Done. Now you can analyse this spectral signature with Analyse Region. The histograms of all bands in the image group will be displayed. To see the matching of this signature in the entire displayed raster image choose Display Matches. You can choose another color here (during your work with several areas), set the standard deviation and leave this menu with Done. Now you are asked if you accept this spectral signature and can give a name in this case.
After that you proceed with the next training area. When you leave the module totally the signatures are saved in the "result signature file".
This file is the input for the "maximum likelyhood" classification. Please refer chapter VII 7.1 for the
use of
$
i.maxlik.The result is a classified image raster layer and a reject threshold map containing the probability of assignment of each pixel to its class.
6.1.3 Semi supervised classification
In a semi supervised classification only some areas will be defined. That can be done using the module
$ i.class
Here you define the boundaries of easy to recognize areas like waterbodies etc. When leaving the module some spectral signatures are stored in the "result signature file". For the rest of the clustering the module
$ i.cluster
will be used. Specify a new name for the final "result signature file" and give the "result signature file" of i.class here as "seed signature". This will be used for initialising the clusters with well known values. So the result will be more accurate than in the unsupervised classification.
The final "result signature" is the input for
$ i.maxlik
Please refer chapter VII 7.1 for more information about the use of this module. The result is a classified image raster layer and a reject threshold map containing the probability of assignment of each pixel to its class.
6.2 Spatial pattern recognition
The spatial pattern recognition takes care of the neighbourhood of pixels. Opposite to the "maximum likelyhood" classifier this "sequential maximum a posteriori" (SMAP) algorithm does not work pixelwise, it is geometrical oriented and uses a matrix window.
Also here the first step is to collect all images in an image group:
$ i.group
You define a group name and mark the images which shall be used for the classification with an "x". Then you have to define a "subgroup" (menue: 5.), give a name to the subgroup and again mark the images. After that you can leave this module.
Now digitize the areas with a digitizer from a map (toposheet etc.). Use either
$ v.digit in combination with
$ v.to.rast
or
$ r.digit
This training map has to be in raster format and will be feeded into the clustering module. Only the specified areas will be used in the classification later on, so all classes you want to have must be digitized. All pixels will be assigned later on, because of this being a supervised classification no additional classes will be added. If you define only less classes the result will be not very accurate.
The resulting clusters calculated from the training areas are the input for the SMAP classification algorithm. The name of the clustering module is
$ i.gensigset
First it asks for the training map, then for the group and the subgroup containing the satellite images. The "subgroup signature file" is comparing to the "result signature file" and the input for the next module
i.smap. Now the cluster will be calculated.The classification itself is performed with
$ i.smap
First group and subgroup have to be given, then the with
i.gensigset created "subgroup signature file". After specifying the output raster filename the classification starts.The result is the classified raster image layer.
7. Enhancement of image resolution using IHS/RGB transformation – image fusion
The method of improving image resolution with IHS/RGB transformation (Intensity, Hue Saturation from/to Red, Green, Blue) is based on the fact, that opposite to the RGB-color system the IHS channels are independent from each other.
For the image resolution enhancement it will be made use of this feature. The satellite images (three channels, mostly red, green and blue channel) are transformed to the IHS system. Then the saturation channel will be replaced with the high resolution channel. After that these three images will be backtransformed to the RGB color-system.
In the example of merging LANDSAT-TM 5 channels (30m resolution) with SPOT panchromatic data (10m resolution) the color information are taken from LANDSAT-TM, the geometrical information from the SPOT.
The actual resolution shall be set to 30m (LANDSAT). To transform the three basic color band images (bands 1,2,3, they are in RGB-color system) into the IHS-system use:
$ i.rgb.his
First these three input channels have to be specified:
1. Red band
2. Green band
3. Blue band
Then the names for the three new output channels in IHS-color system. The resulting three channels are
1. (LANDSAT) Hue
2. (LANDSAT) Intensity
3. (LANDSAT) Saturation
Now you have to change the resolution to the high resolution using
$ g.region res=10m
$ d.erase
The second command is necessary to reset the GRASS Monitor.
Now you backtransform the IHS images to RGB, but you specify the SPOT-panchromatic channel instead of the LANDSAT saturation image. The module's name is
$ i.his.rgb
Specify as input channels
1. (LANDSAT) Hue
2. (LANDSAT) Intensity
3. SPOT panchromatic
The output channels (getting the high 10m resolution) are
1. new red channel
2. new green channel
3. new blue channel
The result are three new LANDSAT images containing the color information of the original images and the high resolution of SPOT. The same can be performed also with IRC-1C etc.
Instead of using the first three bands of LANDSAT you can also use band ratios or other channels.
This overview paper describes some of the GIS capabilities of GRASS GIS software. As to be seen all functions which are required in a hybrid GIS are available. All modules are documented, several specialized papers are available within the GRASS distribution. The user can try most of the GRASS commands on a sample dataset "Rushmore / South Dakota, U.S.A" (SPEARFISH database) which comes along with the distribution and is also stored in Internet.
GRASS can help the user starting from the analog data input maps in form of either digital unreferenced form (e.g. scanned maps), which can be digitized on screen with a mouse digitizer or with an external digitizing board or these data can be imported as digital referenced images or spreadsheet tables. The interface to ARC/INFO's "ungenerate" vectorformat is a flexible way of data integration. All formats are supported: point data, vector line and polygon data as well as raster data.
The strongest points of GRASS software are its raster analysis and image processing facilities. In raster analysis weighted overlaying without limitation of layers up to complex models like erosion modelling can be carried out. The image processing modules contain the standard functions like rectification, supervised and unsupervised classification but also image enhancement with digital filters, fourier transformation, edge detection etc.
The functions are comparing or better than commercial GIS software. But the GRASS software is in public domain, freely available in internet or for less charge on CD-ROM. The modular concept and the fully described programing library allow the user to write modules for his/her own purpose. These modules can make use of the database access functions and predefined standard GIS functions and connect them to own calculations. A lot of examples can be found in the standard GRASS distribution as well as in Internet in external modules with GRASS interface.
Appendix 1: Frequently asked questions and their answers
Question 1: After import of a raster image I cannot display it (I see only an one-color area).
Answer: Please use the module
r.support to "edit the header": Check the coordinates of the imported image. If this doesn't help, useg.region rast=rasterfilename
d.erase
d.rast rasterfilename
With this command you focus the view onto the coordinates of the raster image.
Question 2: While displaying maps using d.vect, d.rast etc., I get the message:
"WARNING: [rasterfile] in mapset [mapset] - in different projection than current region." What does this mean?
Answer: What has happened: You have opened the GRASS Monitor in another location with a different projection before, leaved GRASS without closing the monitor and started GRASS again in a new existing projection. Then you selected (d.mon select=x0) again this monitor which still has the old projection in the display.
Solution: You stop the monitor and start it again.
Question 3: The digitizer
v.digit doesn't seem to work in mouse digitizing mode (what shall I do here?).
Answer: Please read, what is written in the command xterm. If there is a menu displayed, use the keyboard, if there are "button"-commands, use the mouse in the GRASS monitor.
Question 4: After using
r.in.sunrast my satellite images looks somehow unusual and tilted.
Answer: Problably you have given "yes" at the folloing question:
"Don't adjust number of rows [n]:"
Here you should use the default: "no". The way of asking this question in this module is a bit confusing.
Question 5: After converting an raster image with xv I want to import it into GRASS. But the modules
r.in.gif, r.in.tiff etc. does not like my file.
Answer: The main mistake is that the image is stored in the wrong colorformat. That means, GRASS supports the following import formats
· 8 bit: GIF87, TIFF, Sun Raster
· 24 bit: PPM
While saving the image in xv you should look for this bit-format and change it, if necessary. The menu in xv is in the first line: "24/8 bit". Switch here before saving.
Appendix 2: Script programming in GRASS
The following script example demonstrates how to calculate a new raster layer out of old raster layers.
It is a UNIX script, the GRASS modules are called as usually. Parameters can be specified. Please avoid algebraic symbols in filenames for GRASS.
:/bin/sh
echo "Start GRASS before!!!"
echo "Parameter: Call this script with forthnight-number as parameter!"
echo "Example:"
echo " calculate 14 (do it for the forthnight no. 14)"
echo ""
# if you have forgotten the parameters, look into the manual or type in:
# grasscommand help
# example:
# g.remove help
# ---------------------------------------------------------------
r.mapcalc rainfall.rech.$1 = rainfall.$1 - runoff.$1 - et.$1
# The following line is ONE line only!
r.mapcalc total.inflow.$1 = rainfall.rech.$1 + returnflow.$1 + subin.$1 + riverbed.rech.$1
# remove the first layer:
g.remove rainfall.recharge.$1
# go on...
r.mapcalc total.outflow.$1 = extraction.$1 + vet.$1 + subout.$1
r.mapcalc netto.q.$1 = total.inflow.$1 - total.outflow.$1
# remove two layers:
g.remove total.inflow.$1,total.outflow.$1
echo ""
echo "Finished..."
# --------------------------------------------------------------
echo " Display the result:"
# start graphical output
d.mon x0
# display rasterfile:
d.rast netto.q.$1
Appendix 3: GRASS Command Overview
The following command list explains more than 250 GRASS modules. This is only a part of the entire distribution. Many extra modules can be found in the internet and the new GRASS 4.2.1 distribution.
d.3d - Three-dimensional raster map display module.
d.ask - Prompts the user to select a GRASS data base file from among files displayed in a menu on the graphics monitor (for shell-moduleming).
d.colormode - Allows the user to establish whether a map will be displayed using its own color table or the fixed color table of the graphics monitor.
d.colors - Allows the user to interactively change the color table of a raster map layer displayed on the graphics monitor (specify RGB-values).
d.colortable - To display the color table associated with a raster map layer.
d.display - Menu module to display textual, symbolic, raster, vector, and site data in the active frame on the user's graphics monitor.
d.erase - Erases the active display frame on the graphics monitor (use after region settings with g.region).
d.font - Selects text fonts to be used for text display on the graphics monitor.
d.frame - Manages display frames on the graphics monitor.
d.geodesic - Displays a geodesic line, tracing the shortest distance between two geographic points along a great circle, in a longitude/latitude data set.
d.graph - Module for generating and displaying simple graphics to the graphics display monitor (with xy-coordinates of the graphics monitor, refer d.mapgraph).
d.grid - Overlays a user-specified grid in the active display frame on the graphics monitor.
d.his - Produces and displays a raster map layer combining hue, intensity, and saturation (his) values
from user-specified input raster map layers.
d.histogram - Plots a histogram of the cell values (categories) in the form of a pie or bar chart for a user-specified raster file.
d.icons - Displays points, as icons, at user-defined locations in the active display frame on the graphics monitor.
d.labels - To create/edit GRASS label files for display on the graphics monitor.
d.legend - Displays a legend for a raster map layer in the active frame on the graphics monitor.
d.mapgraph - Generates and displays simple graphics on raster map layers drawn in the active graphics monitor display frame.
d.measure - Measures the lengths and areas of features drawn by the user in the active display frame on the graphics monitor.
d.menu - Creates and displays a menu within the active frame on the graphics monitor.
d.mon - To establish and control use of a graphics display monitor.
d.paint.labels - Displays text labels formatted for use with GRASS paint (paint, p.map) output to the active frame on the graphics monitor.
d.points - Displays point graphics in the active frame on the graphics display monitor.
d.profile - Displays profiles of a user-specified raster map layer in the active frame on the user's graphics monitor.
d.rast - Displays and overlays raster map layers in the active display frame on the graphics monitor.
d.rast.arrow - Displays arrows representing aspect directions for a raster map layer in the active display frame on the graphics monitor.
d.rast.edit - Allows users to interactively edit cell category values for a raster map layer displayed on the graphics monitor.
d.rast.num - Displays cell category values contained in raster map layers displayed on the graphics monitor.
d.rast.zoom - Allows users to interactively zoom into regions displayed on the graphics monitor.
d.rgb - Allows multiple maps to be overlaid in the active display frame on the graphics monitor. Better results gives the module i.composite.
d.rhumbline - Displays the rhumbline joining two user-specified points in the active frame on the user's graphics monitor.
d.save - Creates a shell script listing of commands needed to recreate GRASS screen graphics.
d.scale - Displays a map scale and north arrow relevant to a the user's current geographic region setting in the active display frame on the graphics monitor.
d.scale2 - Overlays a bar scale and north arrow for the current geographic region at a user-defined location in the active display frame.
d.sites - Displays site markers in the active display frame on the graphics monitor.
d.text - Draws text in the active display frame on the graphics monitor.
d.title - Outputs a title for a raster map layer in a form suitable for display by d.text.
d.vect - Displays GRASS vector data in the active frame on the graphics monitor.
d.what.rast - Allows the user to interactively query the category contents of multiple raster map layers at user-specified locations within the current geographic region.
d.what.vect - Allows the user to interactively query the category contents of multiple (binary) vector map layers at user-selected locations within the current geographic region.
d.where - Identifies the geographic coordinates associated with point locations in the active frame on the graphics monitor.
d.zoom - Allows the user to change the current geographic region settings interactively, with a mouse.
g.access - Controls user access to the current GRASS mapset.
g.ask - Prompts the user for the names of GRASS data base files.
g.copy - Copies available data files in the user's current mapset search path and location to the appropriate element directories under the user's current mapset.
g.filename - Prints GRASS data base file names.
g.findfile - Searches for GRASS data base files and sets variables for the shell.
g.gisenv - Outputs the user's current GRASS variable settings.
g.list - Lists available GRASS data base files of the specified data type to standard output.
g.manual - Access GRASS reference manual entries.
g.mapsets - Modifies the user's current mapset search path, affecting the user's access to data existing under the other GRASS mapsets in the current location.
g.region - Module to manage the boundary definitions for the geographic region.
g.remove - Remove data base element files from the user's current mapset.
g.rename - To rename data base element files in the user's current mapset.
g.tempfile - Creates a temporary file and prints the file name.
g.version - Outputs the GRASS version number and date.
i.cca - Canonical components analysis (cca) module for image processing.
i.class - An imagery function that generates spectral signatures for an image by allowing the user to outline regions of interest. The resulting signature file can be used as input for i.maxlik or as a seed signature file for i.cluster.
i.cluster - An imagery function that generates spectral signatures for land cover types in an image using a clustering algorithm. The resulting signature file is used as input for i.maxlik. The algorithm is minimum distance.
i.colors - An imagery function that creates colors for imagery groups.
i.composite - An imagery function that creates a color composite image from three band files specified by the user.
i.fft - Fast Fourier Transform (FFT) for image processing.
i.gensig - An imagery function that generates spectral signatures for an image based on a trainging map of already classified areas/pixels. The resulting signature file can be used as input for i.maxlik or as a seed signature file for i.cluster.
i.grey.scale - An imagery function that assigns a histogram contrast stretch grey scale color table to a raster map layer (linear stretch). To stretch the cell values use r.mapcalc.
i.group - An imagery function that creates and edits groups and subgroups of (raster) imagery files.
i.his.rgb - Hue-intensity-saturation (his) to red-green-blue (rgb) raster map color transformation function.
i.ifft - Inverse Fast Fourier Transform (ifft) for image processing.
i.maxlik - An imagery function that classifies the cell spectral reflectances in imagery data based on the spectral signature information generated in i.cluster. The classification is the maximum likelyhood algorithm.
i.median - An interactive imagery function that creates a new raster map layer whose color table is based on the red, green, and blue (RGB) color values present in existing, user-specified imagery group files.
i.ortho.rectify - An imagery function that performs ortho-photo rectification with DEM.
i.pca - Principal components analysis (pca) module for image processing.
i.points - An imagery function that enables the user to mark coordinate system points on an image to be rectified and then input the coordinates of each point for creation of a coordinate transformation
matrix. The transformation matrix is needed as input for the GRASS module i.rectify.
i.rectify - An imagery function that rectifies an image by computing a coordinate transformation for each cell (pixel) in the image using the transformation coefficient matrix created by the GRASS module i.points.
The used transformation is affine.
i.rectify2 - An imagery function that rectifies an image by computing a coordinate transformation for each cell (pixel) in the image using the transformation coefficient matrix created by the GRASS module i.points.
The used transformation is polynomial of definable order.
i.rgb.his - Red-green-blue (rgb) to hue-intensity-saturation (his) function for image processing.
i.tape.mss - An imagery function that extracts Multispectral Scanner (MSS) imagery data from half-inch tape.
i.tape.mss.h - An imagery function that extracts header information from Landsat Multispectral Scanner (MSS) imagery data stored on half-inch tape.
i.tape.other - An imagery function that extracts scanned aerial imagery (NHAP, etc.) and SPOT imagery from magnetic tape or floppy in tar-format.
i.tape.spot - An imagery function that extracts SPOT imagery from half-inch tape.
i.tape.tm - An imagery function that extracts LANDSAT Thematic Mapper (TM) imagery from half-inch magnetic tape.
i.tape.tm.fast - An imagery function that extracts Thematic Mapper (TM) imagery from tape media
i.target - An imagery function that establishes a GRASS target LOCATION for an imagery group.
i.zc - Zero-crossing "edge detection" raster function for image processing.
GRASS data import/processing modules
m.datum.shift - Datum shift module.
m.dem.examine - Provides a terse description of USGS Digital Elevation Model (DEM) data files stored on 1/2-inch magnetic tape.
m.dem.extract - Extracts USGS Digital Elevation Model (DEM) data from 1/2-inch magnetic tape.
m.dem.extract - Provides information about data stored on Digital Elevation Model (DEM) tapes.
m.dmaUSGSread - Extracts digital terrain elevation data (DTED) produced by the Defense Mapping Agency (DMA) but supplied by the USGS (in a different tape format) on 1/2-inch magnetic tape.
m.dted.examine - Provides a terse description of level 1 and 2 digital terrain elevation data (DTED) files produced and distributed by the Defense Mapping Agency (DMA) on 1/2-inch magnetic tapes.
m.dted.extract - Extracts digital terrain elevation data (DTED - levels 1 and 2) produced and supplied by the Defense Mapping Agency (DMA) on 1/2-inch magnetic tapes.
m.eigensystem - Computes eigen values and eigen vectors for square matricies.
m.examine.tape - Provides a description of the files on a 1/2-inch magnetic tape.
m.flip - Flips the orientation of map data.
m.gc2ll - Converts geocentric to geographic coordinates.
m.ll2gc - Converts geographic coordinates to geocentric coordinates.
m.ll2u - Converts geographic (latitude/longitude) coordinates to Universal Transverse Mercator (UTM) coordinates.
m.lulc.USGS - Creates raster map layers from a Composite Theme Grid (CTG) file created by m.lulc.read. m.lulc.read extracts the CTG data from an ASCII landuse/landcover (lulc) CTG format file supplied by the USGS.
m.lulc.read - Extracts Landuse/Landcover data in the ASCII Composite Theme Grid (CTG) data format distributed by the USGS in to a working file for m.lulc.USGS.
m.proj - Calculates conversion coordinates for geographic positions.
m.region.ll - Converts Universal Transverse Mercator (UTM) coordinates falling within the current geographic region from UTM coordinates to geographic (latitude/longitude) coordinates.
m.rot90 - Rotates elevation data extracted by either m.dted.extract or m.dmaUSGSread.
m.tiger.region - Module to extract the boundaries of a geographic region sufficient to encompass all data in a user-specified TIGER file.
m.u2ll - Converts Universal Transverse Mercator (UTM) coordinates to geographic (latitude/longitude) coordinates.
GRASS paint/print output modules
d.label - Creates and places text labels in the active display frame on the graphics monitor.
p.chart - Prints a color chart for the printer currently selected by the user.
p.colors - Allows the user to modify a color table for a raster map layer, assigning colors to the categories in the raster map layer based on printer color numbers (instead of red, green, blue percentages).
p.icons - Allows the user to create and maintain icons which are used by the p.map command to depict sites.
p.labels - Creates and modifies labels for hardcopy maps.
p.map - Command language interface to color hardcopy and graphics monitor output.
p.ppm - Reads binary portable pix-map (ppm) files created by PPM utilities.
p.select - Selects a device (printer) for GRASS hardcopy output. Outputs can also be previewed on the graphics display monitor.
ps.icons - Creates and modifies icons for use with fIps.map fR.
ps.map - Hardcopy PostScript map output utility.
ps.select - Selects a PostScript device for GRASS hardcopy output.
r.average - Finds the average of values in a cover map (binary map) within polygons of a user-specified
base map.
r.basins.fill - Generates a raster map layer showing watershed subbasins.
r.binfer - Bayesian expert system development function.
r.buffer - Creates a raster map layer showing buffer zones surrounding cells that contain non-zero category values.
r.cats - Prints category values and labels associated with user-specified raster map layers.
r.clump - Recategorizes data in a raster map layer by grouping cells that form physically discrete areas into unique categories.
r.coin - Tabulates the mutual occurrence (coincidence) of categories for two raster map layers.
r.colors - Creates/Modifies the color table associated with a raster map layer.
r.combine - Allows category values from several raster map layers to be combined.
r.compress - Compresses and decompresses raster files.
r.contour - Produces a GRASS binary vector map of specified contours from GRASS raster map layer.
r.cost - Outputs a raster map layer showing the cumulative cost of moving between different geographic locations on an input raster map layer whose cell category values represent cost.
r.covar - Outputs a covariance/correlation matrix for user-specified raster map layer(s).
r.cross - Creates a cross product of the category values from multiple raster map layers.
r.describe - Prints terse list of category values found in a raster map layer.
r.digit - An interactive map development module used for digitizing, labeling and creating raster maps.
r.drain - Traces a flow through an elevation model on a raster map layer.
r.grow - Generates an output raster map layer with contiguous areas grown by one cell (pixel).
r.in.ascii - Converts an ASCII text file into a raster map layer.
r.in.ll - Creates a raster file from data in latitude, longitude coordinates.
r.in.poly - Create raster maps from ascii polygon/line data file
r.in.sunrast - Imports a SUN raster file into GRASS
r.infer - Outputs a raster map layer whose category values represent the application of user-specified criteria (rules statements) to other raster map layers' category values.
r.info - Outputs basic information about a user-specified raster map layer.
r.line - Creates a new GRASS vector ( digit ) file by extracting linear features from a thinned raster file.
r.los - Line-of-sight raster analysis module.
r.mapcalc - Powerful raster map layer data calculation tool for overlays etc..
r.mask - Establish/remove the current working mask.
r.median - Finds the median of values in a cover map within polygons of a user-specified base map.
r.mfilter - Raster file matrix filter.
r.mode - Finds the mode of values in a cover map within polygons of a user-specified base map.
r.neighbors - Makes each cell category value a function of the category values assigned to the cells around it, and stores new cell values in an output raster map layer.
r.out.ascii - Converts a raster map layer into an ASCII text file.
r.out.tga - Converts a raster map layer into a Targa format file.
r.patch - Creates a composite raster map layer by using known category values from one (or more) map layer(s) to fill in areas of "no data" in another map layer.
r.poly - Extracts area edges from a raster map layer and converts data to vector format.
r.profile - Plots profiles of a GRASS raster map layer.
r.random - Creates a raster map layer and sites list containing randomly located sites.
r.reclass - Creates a new map layer whose category values are based upon the user's reclassification of categories in an existing raster map layer.
r.report - Reports statistics for raster map layers.
r.resample - GRASS raster map layer data resampling capability (nearest neighbour algorithm).
r.rescale - Rescales the range of category values in a raster map layer.
r.runoff - calculates the runoff in a watershed
r.slope.aspect - Generate raster map layers of slope and aspect from a raster map layer of true elevation values.
r.stats - Generates area statistics for raster map layers.
r.support - Allows the user to create and/or modify raster map layer support files.
r.surf.contour - Surface generation module from rasterized vector contour maps.
r.surf.idw - Surface generation module, from raster data (for latitude/logitude databases with weighted average).
r.surf.idw2 - Surface generation module, using raster data (for projections like UTM etc. with weighted average).
r.surface - Raster surface generation module.
r.thin - Thins non-zero cells that denote linear features in a raster file.
r.transect - Outputs raster map layer category values lying along user-defined transect line(s).
r.volume - Sums cell values within clumps and calculates volumes and centroids of patches or clumps.
r.water.outlet - Generates watershed basin from drainage direction map (from r.watershed) and a set of coordinates representing the outlet point of this watershed (calculates catchment area for this outlet point).
r.watershed - Watershed basin analysis module: calculates watershed boundaries from DEM, drainage map, streams etc.
r.weight - Raster map overlay function.
r.weight.new - Command-line driven raster map overlay function.
r.what - Queries raster map layers on their category values and category labels.
GRASS sites management modules
s.delaunay - Delaunay vector triangulation from sites data.
s.db.rim - RIM data base management/query interface for GRASS.
s.in.ascii - Converts a GRASS site_lists file in ASCII format to binary format.
s.medp - Sites median polish - moving sites onto a grid.
s.menu - Accesses and manipulates GRASS sites data.
s.out.ascii - Converts a GRASS site_lists file in binary format to ASCII format.
s.surf.idw - Surface generation module from sites data (interpolation with weighted average).
s.surf.krig - Surface generation module from sites data (interplation with kriging).
s.surf.tps - Topological analysis and interpolation tool for sites data (spline interpolation with tension).
s.to.rast - Sites to raster conversion.
s.to.vect - Sites to vector conversion.
s.voronoi - Calculation of Thiessen vector polygons from sites data.
v.alabel - Bulk-labels unlabeled area features in a binary GRASS vector file.
v.area - Displays GRASS area and perimeter information for a GRASS vector map layer.
v.cadlabel - Attaches labels to vector contour lines that have been imported to GRASS from DXF format.
v.clean - Cleans out dead lines in GRASS vector files.
v.db.rim - RIM data base management/query interface for GRASS vector maps.
v.digit - A menu-driven, highly interactive map development module used for vector digitizing, editing, labeling and converting vector data to raster format.
v.import - Converts ASCII Digital Line Graph (DLG) files, binary DLG files, and ASCII vector files into binary vector files and creates the needed vector support files.
v.in.arc - Converts data in ARC/INFO format to GRASS's vector format, and gets the input from the user data sored in the current GRASS mapset, subdirecory ./arc.
v.out.arc - Converts data in GRASS format to ARC/INFO vector format, and stores output in the user's current GRASS mapset, subdirecory ./arc.
v.in.ascii - Converts ASCII vector map layers into binary vector map layers.
v.in.dlg - Converts binary DLG (BDLG) file to binary vector (dig) file.
v.in.dxf - DXF format to GRASS vector format conversion module.
v.in.tig.rim - Imports and converts line data in Census Bureau TIGER format to GRASS's vector format using the RIM database system, and stores output in the user's current GRASS mapset.
v.in.transects - import transect data to a GRASS vector map
v.mkgrid - Creates a (binary) GRASS vector map of a user-defined grid.
v.mkquads - Creates a GRASS vector map layer and/or sites list and/or geographic region definition file for a USGS 7.5-minute quadrangle.
v.out.arc - Converts GRASS vector files to ARC/INFO's "Generate" file format.
v.out.ascii - Converts binary vector map layers into ASCII vector map layers.
v.out.dlg - Converts binary GRASS vector data to binary DLG-3 Optional format.
v.out.dxf - GRASS vector format to DXF format conversion module.
v.out.moss - Converts GRASS site, line, or area data into MOSS import format.
v.patch - Creates a new binary vector map layer by combining other binary vector map layers.
v.prune - Prunes points from binary GRASS vector data files.
v.spag - Processes a Spaghetti-digitized vector file.
v.stats - Displays information about a user-specified GRASS vector file.
v.support - Creates GRASS support files for (binary) GRASS vector data.
v.to.rast - Converts a map layer in (binary) GRASS vector format to GRASS raster format.
v.to.sites - Converts a GRASS vector file to a GRASS site_lists file.
v.transform - Transforms an ASCII vector map layer from one coordinate system into another coordinate system.
v.trim - Trims small spurs, and removes excessive nodes from a GRASS vector ( digit ) file.
blend.sh - Combines the color components of two raster map layers and creates three new raster map layers representing the red, green, and blue (rgb) components of the resultant image.
bug.report.sh - GRASS macro that allows users to input bug report information.
dcorrelate.sh - Graphically displays the correlation among from two to four raster map layers in the active frame on the graphics monitor.
demo.sh - Interactive graphic tour of GRASS module functions, using display frames on the graphics monitor.
grass.logo.sh - Displays a GRASS/Army Corps of Engineers logo in the active display frame on the graphics monitor.
hsv.rgb.sh - A GRASS macro, transforming pixel hue, saturation, and v (hsv) values to red, green, and blue (rgb) values.
old.cmd.sh - GRASS macro that outputs the 4.0 module name assigned to any given 3.0 version command.
rgb.hsv.sh - A GRASS macro, transforming pixel red, green, and blue values to hue, saturation, and v (hsv) values.
shade.rel.sh - Builds a shaded relief map given a raster map layer of elevation and the sun's azimuth and altitude above the horizon.
show.color.sh - Displays and names available primary colors used by GRASS modules, in frames on the graphics monitor.
show.fonts.sh - Displays and names available font types in the active display frame on the graphics monitor.
slide.show.sh - Displays a series of raster map layers existing in the user's current mapset search path on the graphics monitor.
split.sh - GRASS macro that creates and makes current two display frames on the graphics monitor.
start.man.sh - GRASS shell script macro that builds a template for GRASS manual entries.
tig.rim.sh - Generates various vector map layers from a RIM/TIGER database.
These descriptions were taken from the GRASS-help system and slightly changed.
Appendix 4: Linux Hardware compatibility list
(List from 6/96, this list is monthly expanded)
1. Main board
Architecture: ISA, EISA, VLB, PCI,
(Microchannel-architecture is not supported)
2. Processors
All Intel 386, 486, 586 or 686 and compatible. Newer Cyrix 6x86 will be supported as 486.
Clones from AMD, Cyrix unt TI run without problems.
3. RAM
At least 12 MB, for X-Window at least 16 MB recommended. More RAM is better than a faster
processor.
4. Hard-drive controller
(a) All AT-Bus-Controller (also "Enhanced IDE-Controller"), ST506-compatible
Controller (e.g. Adaptec ESDI)
(b) SCSI: AdvanSys (all modeltypes), AM53 / 79C974, AMI Fast Disk VLB/EISA (with BusLogic
driver), Adaptec AVA-1505/1515 (ISA) (with 152x driver), Adaptec AHA-1510/152x (ISA), Adaptec AHA-154x (ISA) (all modeltypes), Adaptec AHA-174x (EISA) (in Enhanced Mode), Adaptec AHA-274x (EISA) / 284x (VLB) (AIC-7770), Adaptec AHA-294x (PCI), Adaptec AHA-2920 (with Future Domain driver), AL-500 SCSI Always IN2000, BusLogic (all modeltypes except FlashPoint LT), DPT Smartcache (EATA) (ISA/EISA/PCI), DTC 3180 / 3280, DTC 329x (EISA) (Adaptec 154x compatible), Future Domain TMC-16x0 and TMC-3260 (PCI), Future Domain TMC-8xx and TMC-950, IOMEGA PC2/2B NCR 53C400
(Trantor T 130B), NCR 53C406a, Acculogic ISApport NCR 53c7x0, 53c8x0 (PCI), Pro Audio Spectrum 16 SCSI (ISA), Qlogic / Control Concepts SCSI/IDE (FAS408) - ISA/VLB/PCMCIA, SCSI INN 2000, Seagate ST-01/ST-02 (ISA), SoundBlaster 16 SCSI-2 (Adaptec 152x) (ISA), Trantor T128/T128F/T228 (ISA), UltraStor 14F (ISA), 24F (EISA), 34F (VLB), Western Digital WD7000 SCSI
5. Graphic adapter
(a) Adapter with S3-accelerator:
732 (Trio32), 764 (Trio64), 801/805 AT&T 20C490 (or similar) RAMDAC 805 VLB, S3 GENDAC (RAMDAC + Clock Synthesizer), 801/805 AT&T 20C490 RAMDAC ICD2061A Clockchip, 805 Diamond SS2410 RAMDAC ICD2061A Clockchip, 801/805 Chrontel 8391 Clockchip/RAMDAC, 928 AT&T 20C490 RAMDAC, 928 Sierra SC15025 RAMDAC ICD2061A Clockchip, 928 Bt9485 RAMDAC ICD2061A Clockchip, 928 Bt485 RAMDAC SC11412 Clockchip, 928 Bt485 RAMDAC ICD2061A Clockchip, 928 Ti3020 RAMDAC ICD2061A Clockchip, 864 AT&T20c498 ICS2494 Clockchip, 864 AT&T20c498 or STG1700 RAMDAC ICD2061A or ICS9161 Clockchip, 864 STG1700 RAMDAC ICD2061A Clockchip, 864 20C498 or 21C498 RAMDAC ICS2595 Clockchip, 864 S3 86C716 SDAC RAMDAC and Clockchip, 864 ICS5342 RAMDAC and Clockchip, 864 AT&T21C498-13 RAMDAC ICD2061A Clockchip, 868 S3 Gendac, 964 AT&T
20C505 RAMDAC ICD2061A Clockchip, 964 Bt485 RAMDAC ICD2061A Clockchip, 964 Bt9485 RAMDAC ICS9161a Clockchip, 964 Ti3020 RAMDAC ICD2061A Clockchip, 964 Ti3025 RAMDAC Ti3025 Clockchip, 968 IBM RAMDAC 514/24/25/28, 968 Ti3026 RAMDAC Ti3026 Clockchip, 968 IBM RAMDAC 514/24/25/28
(b) Adapter without S3-accelerator:
#9 FX Motion 531, #9 FX Motion 771, #9 GXE Level 10/ 11/ 12/ 14/ 16, #9 GXE64, #9 GXE64 - PCI, #9 GXE64 Pro, #9 GXE64 Pro PCI, #9 GXE64 Pro VLB, #9 GXE64 Trio64, 928Movie, ASUS Video Magic PCI V864, ASUS Video Magic PCI VT64, Actix GE32, Actix GE32+ 2 MB, Actix GE32i, Actix GE64, Actix Ultra, Dell S3-805, Diamond Stealth 24, Diamond Stealth 64, Diamond Stealth 64 DRAM (only some adapters), Diamond Stealth 64 DRAM SE, Diamond Stealth 64 DRAM with S3-SDAC, Diamond Stealth 64 DRAM with S3-Trio64, Diamond Stealth 64 VIDEO VRAM, Diamond Stealth 64 VRAM, Diamond Stealth Pro, Diamond Stealth VRAM, ELSA Winner 1000 AVI, ELSA Winner 1000 ISA,
ELSA Winner 1000 ISA/EISA (Twinbus), ELSA Winner 1000 PRO, ELSA Winner 1000 PRO VLB, ELSA Winner 1000 PRO with S3-SDAC ELSA Winner 1000 PRO with STG1700 or AT&T RAMDAC, ELSA Winner 1000 VL, ELSA Winner 2000, ELSA Winner 2000 ISA/EISA, ELSA Winner 2000 PRO/X2, ELSA Winner 2000 PRO/X4, Genoa Phantom 64i with S3-SDAC, Genoa VideoBlitz III AV, Hercules Graphics Terminator 64, Hercules Graphics Terminator Pro 64, JAX 8241, Miro Crystal 8S, Miro Crystal 10SD, Miro Crystal 16S, Miro Crystal 20SD, Miro Crystal 20SV, Miro Crystal 40SV, Orchid Fahrenheit 1280, Orchid Fahrenheit 1280+ VLB, Orchid Fahrenheit VA, SPEA/V7 Mercury 2MB VL,
SPEA/V7 Mercury P64, SPEA/V7 Mirage, SPEA/V7 Mirage P64, STB Pegasus, STB Powergraph X-24 S3, STB Velocity 64, Spider Tarantula 644, VL-414, VidTech FastMax P204, VideoMagic PCI V8644
Supported Clock-Chips:
DCS2824-0 (Diamond, ICD2061A comp.), Clockchip "dcs2824" ICD2061A, Clockchip "icd2061a" ICS2595, Clockchip "ics2595" ICS5300 GENDAC (86c708 compatible), Clockchip "ics5300" ICS5342 GENDAC, Clockchip "ics5342, Clockchip ICS9161A (ICD2061A compatible), Clockchip "ics9161a" S3 86c708 GENDAC, Clockchip "s3gendac" S3 86c716 SDAC, Clockchip "s3 sdac" Sierra SC11412, Clockchip "sc11412" TI3025, Clockchip "ti3025" TI3026, Clockchip "ti3026"
(c) Adapter with ET4000/W32/W32p/W32i:
Cardex Challenger (Pro), Cardex Cobra, DFI-WG5000, Genoa 8900 Phantom 32i, Hercules Dynawithe Power, Hercules Dynawithe Pro, LeadTek WinFast S200, STB LightSpeed, Sigma Concorde, TechWorks Thunderbolt, ViewTop PCI
(d) Adapter with Weitek P9000:
Diamond Viper (PCI & VLB), Orchid P9000
(e) Adapter with IIT AGX: Chips:
AGX-016, AGX-015 and AGX-014, Hercules Graphite HG210, Hercules Graphite Power, Hercules Graphite Pro, Orchid Celsius (AT&T RAMDAC), Orchid Celsius (Sierra RAMDAC), Spider Black Widow, Spider Black Widow Plus, XGA-1 (ISA Bus), XGA-2 (ISA Bus)
(e) SVGA (fast):
Cirrus Logic 542x, 5430 (16 bpp), 543x (16/32 bpp),
62x5 adapters:
ALG-5434, Actix ProStar, Actix ProStar 64, Cirrus Logic ..., Diamond SpeedStar 64, Diamond SpeedStar Pro, Diamond SpeedStar Pro SE, Genoa 8500VL, Intel 5430, Orchid Kelvin 64 ISA/VLB, STB Horizon, STB Nitro, Spider VLB Plus VI720
(f) Oak OTI-087 adapters:
Paradise Accelerator Value
(g) ARK 1000PV/2000PV adapters:
Hercules Stingray Pro, Hercules Stingray 64
(h) SVGA (normal):
ATI VGA Wonder series, Avance Logic AL2101/2228/2301/2302/2308/2401, Hercules Stingray, SPEA/V7 Mirage VEGA Plus, Chips & Technologies 65520/65530/65540/65545, Cirrus Logic 5420/5422/6420/6440, Acumos AVGA3, DFI-WG1000, Cirrus Logic GD62xx (Laptop), Cirrus Logic GD64xx (Laptop), Compaq AVGA, Genoa GVGA MCGA (320x200), MX MX68000/MX68010, NCR 77C22/
77C22E/ 77C22E+, Oak OTI-067/ OTI-077, Trident TVGA8800, TVGA8900, TVGA9xxx (not TGUI9660), Tseng ET3000, ET4000AX, ET4000, Diamond SpeedStar (Plus), Video 7 / Headland Technologies HT216-32, Western Digital/Paradise PVGA1, D90C00/10/11/24/30/31/33, DFI-WG6000, Diamond SpeedStar 24X
(i) VGA16 colors:
Nearly all (some Cirrus Logic work only with SVGA-Server)
(j) Monochrome:
Hercules mono, Hyandai HGC-128, Sigma LaserView PLUS, VGA mono
6. Network
3Com 3C501/ 3C503/ 3C505/ 3C507/ 3C509 (ISA)/ 3C579 (EISA)/ 3Com 3c590 Serie
(592/595/597) "Vortex", Ansel Communications AC3200 EISA, Apricot Xen-II, AMD LANCE
(79C960), Allied Telesis AT1700, Arcnet (all adapters), AT-LAN-TEC / RealTek, Cabletron
E21xx, DEC DEPCA and EtherWORKS, Digital DE425/434/435/450/500 / DEC21040 (PCI), D-
Link 600/620 Pocket Adapter, Fujitsu FMV-181/182, Gracilis PackeTwin, HP PCLAN (27245 and
27xxx Serie), HP PCLAN PLUS (27247B and 27252A), IBM Token Ring (Tropic Chipsatz), ICL
EtherTeam 16i/32, Intel EtherExpress 16/Pro, NE2000, NE1000, Ottawa PI and PI2, PCnet-
ISA/PCI (AT1500/ HP J2405A/ NE1500/NE2100), PureData PDUC8028/ PDI8023, Racal-Interlan
NI5210 (i82586 Ethernet chip), Racal-Interlan NI6510 (am7990 lance chip), SMC Ultra Schneider
& Koch G16, Western Digital WD80x3, WaveLAN AT&T GIS and NCR, WaveLAN Zenith Z-
Note
7. Mouse (best choice is three button mouse)
Serielle Mouse (Microsoft/ Logitech/ Mouseman/ Mousesystems), Logitech Busmaus,
Mousesystems (3 buttons), PS/2 Busmouse, C&T 82C710 mouse port, Microsoft Busmouse, ATIXL
Busmouse
8. CDROM
All SCSI-drives (connected to supported SCSI-Controller), Aztech CDA268, Orchid CDS-3110,
TXC, Okano/Wearnes CDD-110, EIDE (ATAPI) CD-ROM drives (all), Goldstar R-420, Mitsumi
Lu005, FX001, FX001D drives at Mozart Soundcard, Optics Storage DOLPHIN 8000AT, Philips
CM 206, Sanyo CDR-H94A, Sony CDU-31A/ CDU-33A/ CDU-531/ CDU-535/ CDU-541,
Soundblaster CD-ROMs (Matsushita, Panasonic, Kotobuki), Pro Audio Spectrum CD-ROMs
9. Tape Streamer
All SCSI-Streamer (connected to supported SCSI-Controller), EIDE (ATAPI) Streamer, diverse
Floppystreamer (IOMega etc.).
10. Soundkarten
6850 UART MIDI, ATI Stereo F/X (SB compatible), Adlib Audio Excell DSP16, Cardinal
DSP16, Crystal CS4232 based adapters, ECHO-PSS (Orchid SW32, Cardinal DSP16, etc),
Ensoniq SoundScape (boot DOS to init card), Gravis Ultrasound, Gravis Ultrasound ACE, Gravis
Ultrasound 16-bit, Gravis Ultrasound MAX, Logitech SoundMan Games (SBPro, 44kHz Stereo
Support), Logitech SoundMan Wave (SBPro/MPU-401) (OPL4), Logitech SoundMan 16 (PAS-16
compatible), Microsoft Sound System (AD1848), MAD16 and Mozart based adapters, MPU-401
MIDI, MediaTriX AudioTriX Pro, Media Vision Premium 3D (Jazz16) (SBPro compatible),
Media Vision Pro Sonic 16 (Jazz), Media Vision Pro Audio Spectrum-16, Orchid SW32 Pro
Audio Spectrum (NOT in combination with Adaptec AHA 1542, because of conflicts with DMA-
access), Pro Sonic 16, SoundBlaster, SoundBlaster Pro, SoundBlaster 16/ASP/MCD/SCSI-2,
Sound Galaxy NX Pro ThanderBoard (SB compatible), Turtle Beach Maui and Tropez,
WaveBlaster (and other SB16 compatible), Windows Sound System, Yamaha FM Synthesizer
(OPL2/ OPL3/ OPL4)
Appendix 5: Internet sites for GRASS
The servers available in Internet can be distinguished into software and information server. The software server are called "FTP-server", the information server "WWW-Server". The WWW-server are userfriendly and contain mostly links to software sources.
The GRASS source code itself is stored on the following servers:
The main server of GRASS Research Group:
http://www.baylor.edu/~grass/ (U.S.A.)
GRASS 4.2.1 Server:
http://www.laum.uni-hannover.de/iln/grass/grass42/ (Germany)
Mirrors of GRASS 4.1:
ftp://ftp.informatik.uni-kiel.de/pub/packages/grass/ (Germany, sometimes not
available)
ftp://ftp.funet.fi/pub/graphics/packages/grass/ (Finland)
ftp://ftp.funet.fi/pub/graphics/packages/grass-incoming/ (Finland)
For some UNIX-platforms precompiled binaries can be found at Baylor’s server: SUN, Linux etc.
Information to modules, handbooks (also german language), projects you can find at Hannover University, Germany:
http://www.laum.uni-hannover.de/iln/grass/
http://www.geog.uni-hannover.de/phygeo/grass/
The discussion newsgroups are:
news:info.grass.programmer
news:info.grass.user
These discussion groups can also be accessed through a mailing list. Information are available at the main GRASS WWW-server at CERL (links stored in Hannover-pages).
A new GRASS module page for Linux (precompiled binaries with source code) is accessible at
http://www.laum.uni-hannover.de/iln/grass/linuxsoftware/linux-binaries/
From these Hannover servers there are links to several other WWW-servers.
The Software package with documentation is also available an CDROM. Information you find on the GRASS 4.2.1 Server
http://www.laum.uni-hannover.de/iln/grass/grass42/
especially on the page
http://www.laum.uni-hannover.de/iln/grass/grass42/cdrom.html
Albrecht, J. (1992): GTZ-handbook GRASS. Vechta, Germany.
Available through internet at:
http://www.laum.uni-hannover.de/iln/grass/
Byars, B.W., Clamons, S.F. (1997): GRASS 4.2 Reference Manual. Waco, U.S.A.
Available through internet at:
http://www.baylor.edu/~grass/
Fox, J. (1989): RIM database system. Installers manual. University of Washington.
Stored on the GRASS CDROM and Internet.
Fox, J. (1989): RIM database system. Users manual. University of Washington.
Stored on the GRASS CDROM and Internet.
Harmon, V., Shapiro, M. (1992): GRASS tutorial: Image processing. CERL, Champaign, Illinois.
Stored on the GRASS CDROM and Internet.
Jensen, J. R. (1996): Introductory Digital Image Processing: A remote sensing perspective.
Second Edition. New Jersey.
Larson, M., Shapiro, M., Tweddale, S. (1991): Performing map calculations on GRASS data: r.mapcalc programming tutorial. CERL, Champaign, Illinois.
Available through internet at:
http://www.laum.uni-hannover.de/iln/grass/
Lillesand, Th. M., Kiefer, R. W. (1994): Remote sensing and image interpretation. New York.
Neidig, C.A., Gerdes, D., Kos, Ch. (1991): GRASS 4.0 map digitizing manual: v.digit. CERL, Champaign, Illinois.
Stored on the GRASS CDROM and Internet.
Neteler, M. (1998): Das GRASS Handbuch. Ein problemorientierter Leitfaden. 3. Auflage, Han
nover, Germany.
Pre-version: http://www.laum.uni-hannover.de/iln/grass/handbuch/
Shapiro, M., Westerveld, J. (1991):
GRASS programmer's manual. CERL, Champaign, Illinois.Stored in Internet.
Shapiro, M., Westerveld, J. (1991): GRASS user's manual. CERL, Champaign, Illinois.
Stored in Internet.
Shapiro, M., Westerveld, J. (1992): r.mapcalc. An algebra for GIS and image processing. CERL, Champaign, Illinois.
Stored in Internet.
Several more specific descriptions are available in Internet or on the GRASS CD-ROM.