Note: This document is for an older version of GRASS GIS that has been discontinued. You should upgrade, and read the current manual page.

NAME
i.vi - Calculates different types of vegetation indices.Uses red and nir bands mostly, and some indices require additional bands.
KEYWORDS
imagery, vegetation index, biophysical parameters, NDVISYNOPSIS
Flags:
- --overwrite
- Allow output files to overwrite existing files
- --help
- Print usage summary
- --verbose
- Verbose module output
- --quiet
- Quiet module output
- --ui
- Force launching GUI dialog
Parameters:
- output=name [required]
- Name for output raster map
- viname=type [required]
- Type of vegetation index
- Options: arvi, ci, dvi, evi, evi2, gvi, gari, gemi, ipvi, msavi, msavi2, ndvi, ndwi, pvi, savi, sr, vari, wdvi
- Default: ndvi
- arvi: Atmospherically Resistant Vegetation Index
- ci: Crust Index
- dvi: Difference Vegetation Index
- evi: Enhanced Vegetation Index
- evi2: Enhanced Vegetation Index 2
- gvi: Green Vegetation Index
- gari: Green Atmospherically Resistant Vegetation Index
- gemi: Global Environmental Monitoring Index
- ipvi: Infrared Percentage Vegetation Index
- msavi: Modified Soil Adjusted Vegetation Index
- msavi2: second Modified Soil Adjusted Vegetation Index
- ndvi: Normalized Difference Vegetation Index
- ndwi: Normalized Difference Water Index
- pvi: Perpendicular Vegetation Index
- savi: Soil Adjusted Vegetation Index
- sr: Simple Ratio
- vari: Visible Atmospherically Resistant Index
- wdvi: Weighted Difference Vegetation Index
- red=name
- Name of input red channel surface reflectance map
- Range: [0.0;1.0]
- nir=name
- Name of input nir channel surface reflectance map
- Range: [0.0;1.0]
- green=name
- Name of input green channel surface reflectance map
- Range: [0.0;1.0]
- blue=name
- Name of input blue channel surface reflectance map
- Range: [0.0;1.0]
- band5=name
- Name of input 5th channel surface reflectance map
- Range: [0.0;1.0]
- band7=name
- Name of input 7th channel surface reflectance map
- Range: [0.0;1.0]
- soil_line_slope=float
- Value of the slope of the soil line (MSAVI and PVI only)
- soil_line_intercept=float
- Value of the intercept of the soil line (MSAVI only)
- soil_noise_reduction=float
- Value of the factor of reduction of soil noise (MSAVI only)
- storage_bit=integer
- Maximum bits for digital numbers
- If data is in Digital Numbers (i.e. integer type), give the max bits (i.e. 8 for Landsat -> [0-255])
- Options: 7, 8, 10, 16
- Default: 8
Table of contents
DESCRIPTION
i.vi calculates vegetation indices based on biophysical parameters.- ARVI: Atmospherically Resistant Vegetation Index
- CI: Crust Index
- DVI: Difference Vegetation Index
- EVI: Enhanced Vegetation Index
- EVI2: Enhanced Vegetation Index 2
- GARI: Green atmospherically resistant vegetation index
- GEMI: Global Environmental Monitoring Index
- GVI: Green Vegetation Index
- IPVI: Infrared Percentage Vegetation Index
- MSAVI2: second Modified Soil Adjusted Vegetation Index
- MSAVI: Modified Soil Adjusted Vegetation Index
- NDVI: Normalized Difference Vegetation Index
- NDWI: Normalized Difference Water Index
- PVI: Perpendicular Vegetation Index
- RVI: ratio vegetation index
- SAVI: Soil Adjusted Vegetation Index
- SR: Simple Vegetation ratio
- WDVI: Weighted Difference Vegetation Index
Background for users new to remote sensing
Vegetation Indices are often considered the entry point of remote sensing for Earth land monitoring. They are suffering from their success, in terms that often people tend to harvest satellite images from online sources and use them directly in this module.
From Digital number to Radiance:
Satellite imagery is commonly stored in Digital Number (DN) for
storage purposes; e.g., Landsat5 data is stored in 8bit values
(ranging from 0 to 255), other satellites maybe stored in 10 or 16
bits. If the data is provided in DN, this implies that this imagery
is "uncorrected". What this means is that the image is what the
satellite sees at its position and altitude in space (stored in DN).
This is not the signal at ground yet. We call this data at-satellite
or at-sensor. Encoded in the 8bits (or more) is the amount of energy
sensed by the sensor inside the satellite platform. This energy is
called radiance-at-sensor. Generally, satellites image providers
encode the radiance-at-sensor into 8bit (or more) through an affine
transform equation (y=ax+b). In case of using Landsat imagery, look
at the i.landsat.toar for an easy way to transform DN to
radiance-at-sensor. If using Aster data, try the i.aster.toar
module.
From Radiance to Reflectance:
Finally, once having obtained the radiance at sensor values, still
the atmosphere is between sensor and Earth's surface. This fact
needs to be corrected to account for the atmospheric interaction
with the sun energy that the vegetation reflects back into space.
This can be done in two ways for Landsat. The simple way is through
i.landsat.toar, use e.g. the DOS correction. The more
accurate way is by using i.atcorr (which works for many
satellite sensors). Once the atmospheric correction has been applied
to the satellite data, data vales are called surface reflectance.
Surface reflectance is ranging from 0.0 to 1.0 theoretically (and
absolutely). This level of data correction is the proper level of
correction to use with i.vi.
Vegetation Indices
ARVI: Atmospheric Resistant Vegetation IndexARVI is resistant to atmospheric effects (in comparison to the NDVI) and is accomplished by a self correcting process for the atmospheric effect in the red channel, using the difference in the radiance between the blue and the red channels (Kaufman and Tanre 1996).
arvi( redchan, nirchan, bluechan )
ARVI = (nirchan - (2.0*redchan - bluechan)) /
( nirchan + (2.0*redchan - bluechan))
Advantage is taken of a unique spectral feature of soil biogenic crust containing cyanobacteria. It has been shown that the special phycobilin pigment in cyanobacteria contributes in producing a relatively higher reflectance in the blue spectral region than the same type of substrate without the biogenic crust. The spectral crust index (CI) is based on the normalized difference between the RED and the BLUE spectral values (Karnieli, 1997, DOI: 10.1080/014311697218368).
ci ( bluechan, redchan )
CI = 1 - (redchan - bluechan) /
(redchan + bluechan)
DVI: Difference Vegetation Index
dvi( redchan, nirchan ) DVI = ( nirchan - redchan )
EVI: Enhanced Vegetation Index
The enhanced vegetation index (EVI) is an optimized index designed to enhance the vegetation signal with improved sensitivity in high biomass regions and improved vegetation monitoring through a de-coupling of the canopy background signal and a reduction in atmosphere influences (Huete A.R., Liu H.Q., Batchily K., van Leeuwen W. (1997). A comparison of vegetation indices global set of TM images for EOS-MODIS. Remote Sensing of Environment, 59:440-451).
evi( bluechan, redchan, nirchan )
EVI = 2.5 * ( nirchan - redchan ) /
( nirchan + 6.0 * redchan - 7.5 * bluechan + 1.0 )
EVI2: Enhanced Vegetation Index 2
A 2-band EVI (EVI2), without a blue band, which has the best similarity with the 3-band EVI, particularly when atmospheric effects are insignificant and data quality is good (Zhangyan Jiang ; Alfredo R. Huete ; Youngwook Kim and Kamel Didan 2-band enhanced vegetation index without a blue band and its application to AVHRR data. Proc. SPIE 6679, Remote Sensing and Modeling of Ecosystems for Sustainability IV, 667905 (october 09, 2007) doi:10.1117/12.734933).
evi2( redchan, nirchan )
EVI2 = 2.5 * ( nirchan - redchan ) /
( nirchan + 2.4 * redchan + 1.0 )
GARI: green atmospherically resistant vegetation index
The formula was actually defined: Gitelson, Anatoly A.; Kaufman, Yoram J.; Merzlyak, Mark N. (1996) Use of a green channel in remote sensing of global vegetation from EOS- MODIS, Remote Sensing of Environment 58 (3), 289-298. doi:10.1016/s0034-4257(96)00072-7
gari( redchan, nirchan, bluechan, greenchan )
GARI = ( nirchan - (greenchan - (bluechan - redchan))) /
( nirchan + (greenchan - (bluechan - redchan)))
GEMI: Global Environmental Monitoring Index
gemi( redchan, nirchan )
GEMI = (( (2*((nirchan * nirchan)-(redchan * redchan)) +
1.5*nirchan+0.5*redchan) / (nirchan + redchan + 0.5)) *
(1 - 0.25 * (2*((nirchan * nirchan)-(redchan * redchan)) +
1.5*nirchan+0.5*redchan) / (nirchan + redchan + 0.5))) -
( (redchan - 0.125) / (1 - redchan))
GVI: Green Vegetation Index
gvi( bluechan, greenchan, redchan, nirchan, chan5chan, chan7chan)
GVI = ( -0.2848 * bluechan - 0.2435 * greenchan -
0.5436 * redchan + 0.7243 * nirchan + 0.0840 * chan5chan-
0.1800 * chan7chan)
IPVI: Infrared Percentage Vegetation Index
ipvi( redchan, nirchan ) IPVI = nirchan/(nirchan+redchan)
MSAVI2: second Modified Soil Adjusted Vegetation Index
msavi2( redchan, nirchan ) MSAVI2 = (1/2)*(2*NIR+1-sqrt((2*NIR+1)^2-8*(NIR-red)))
MSAVI: Modified Soil Adjusted Vegetation Index
msavi( redchan, nirchan ) MSAVI = s(NIR-s*red-a) / (a*NIR+red-a*s+X*(1+s*s))
NDVI: Normalized Difference Vegetation Index
ndvi( redchan, nirchan ) Satellite specific band numbers ([NIR, Red]): MSS Bands = [ 7, 5] TM1-5,7 Bands = [ 4, 3] TM8 Bands = [ 5, 4] Sentinel-2 Bands = [ 8, 4] AVHRR Bands = [ 2, 1] SPOT XS Bands = [ 3, 2] AVIRIS Bands = [51, 29] NDVI = (NIR - Red) / (NIR + Red)
NDWI: Normalized Difference Water Index (after McFeeters, 1996)
This index is suitable to detect water bodies.
ndwi( greenchan, nirchan ) NDWI = (green - NIR) / (green + NIR)
The water content of leaves can be estimated with another NDWI (after Gao, 1996):
ndwi( greenchan, nirchan ) NDWI = (NIR - SWIR) / (NIR + SWIR)
PVI: Perpendicular Vegetation Index
pvi( redchan, nirchan, soil_line_slope ) PVI = sin(a)NIR-cos(a)red
SAVI: Soil Adjusted Vegetation Index
savi( redchan, nirchan ) SAVI = ((1.0+0.5)*(nirchan - redchan)) / (nirchan + redchan +0.5)
SR: Simple Vegetation ratio
sr( redchan, nirchan ) SR = (nirchan/redchan)
VARI: Visible Atmospherically Resistant Index VARI was designed to introduce an atmospheric self-correction (Gitelson A.A., Kaufman Y.J., Stark R., Rundquist D., 2002. Novel algorithms for estimation of vegetation fraction Remote Sensing of Environment (80), pp76-87.)
vari = ( bluechan, greenchan, redchan ) VARI = (green - red ) / (green + red - blue)
WDVI: Weighted Difference Vegetation Index
wdvi( redchan, nirchan, soil_line_weight ) WDVI = nirchan - a * redchan if(soil_weight_line == None): a = 1.0 #slope of soil line
EXAMPLES
Calculation of DVI
The calculation of DVI from the reflectance values is done as follows:g.region raster=band.1 -p i.vi blue=band.1 red=band.3 nir=band.4 viname=dvi output=dvi r.univar -e dvi
Calculation of EVI
The calculation of EVI from the reflectance values is done as follows:g.region raster=band.1 -p i.vi blue=band.1 red=band.3 nir=band.4 viname=evi output=evi r.univar -e evi
Calculation of EVI2
The calculation of EVI2 from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=evi2 output=evi2 r.univar -e evi2
Calculation of GARI
The calculation of GARI from the reflectance values is done as follows:g.region raster=band.1 -p i.vi blue=band.1 green=band.2 red=band.3 nir=band.4 viname=gari output=gari r.univar -e gari
Calculation of GEMI
The calculation of GEMI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=gemi output=gemi r.univar -e gemi
Calculation of GVI
The calculation of GVI (Green Vegetation Index - Tasseled Cap) from the reflectance values is done as follows:g.region raster=band.3 -p # assuming Landsat-7 i.vi blue=band.1 green=band.2 red=band.3 nir=band.4 band5=band.5 band7=band.7 viname=gvi output=gvi r.univar -e gvi
Calculation of IPVI
The calculation of IPVI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=ipvi output=ipvi r.univar -e ipvi
Calculation of MSAVI
The calculation of MSAVI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=msavi output=msavi r.univar -e msavi
Calculation of NDVI
The calculation of NDVI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=ndvi output=ndvi r.univar -e ndvi
Calculation of NDWI
The calculation of NDWI from the reflectance values is done as follows:g.region raster=band.2 -p i.vi green=band.2 nir=band.4 viname=ndwi output=ndwi r.colors ndwi color=byg -n r.univar -e ndwi
Calculation of PVI
The calculation of PVI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 soil_line_slope=0.45 viname=pvi output=pvi r.univar -e pvi
Calculation of SAVI
The calculation of SAVI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=savi output=savi r.univar -e savi
Calculation of SR
The calculation of SR from the reflectance values is done as follows:g.region raster=band.3 -p i.vi red=band.3 nir=band.4 viname=sr output=sr r.univar -e sr
Calculation of VARI
The calculation of VARI from the reflectance values is done as follows:g.region raster=band.3 -p i.vi blue=band.2 green=band.3 red=band.4 viname=vari output=vari r.univar -e vari
Landsat TM7 example
The following examples are based on a LANDSAT TM7 scene included in the North Carolina sample dataset.Preparation: DN to reflectance
As a first step, the original DN (digital number) pixel values must be converted to reflectance using i.landsat.toar. To do so, we make a copy (or rename the channels) to match i.landsat.toar's input scheme:
g.copy raster=lsat7_2002_10,lsat7_2002.1 g.copy raster=lsat7_2002_20,lsat7_2002.2 g.copy raster=lsat7_2002_30,lsat7_2002.3 g.copy raster=lsat7_2002_40,lsat7_2002.4 g.copy raster=lsat7_2002_50,lsat7_2002.5 g.copy raster=lsat7_2002_61,lsat7_2002.61 g.copy raster=lsat7_2002_62,lsat7_2002.62 g.copy raster=lsat7_2002_70,lsat7_2002.7 g.copy raster=lsat7_2002_80,lsat7_2002.8
Calculation of reflectance values from DN using DOS1 (metadata obtained from p016r035_7x20020524.met.gz):
i.landsat.toar input=lsat7_2002. output=lsat7_2002_toar. sensor=tm7 \ method=dos1 date=2002-05-24 sun_elevation=64.7730999 \ product_date=2004-02-12 gain=HHHLHLHHL
Calculation of NDVI
The calculation of NDVI from the reflectance values is done as follows:
g.region raster=lsat7_2002_toar.3 -p
i.vi red=lsat7_2002_toar.3 nir=lsat7_2002_toar.4 viname=ndvi \
output=lsat7_2002.ndvi
r.colors lsat7_2002.ndvi color=ndvi
d.mon wx0
d.rast.leg lsat7_2002.ndvi
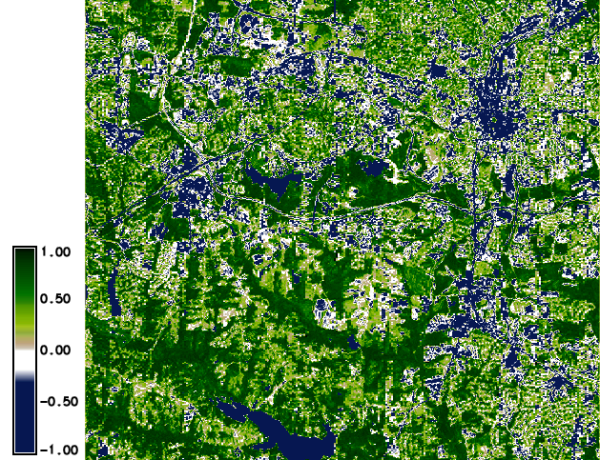
North Carolina dataset: NDVI
Calculation of ARVI
The calculation of ARVI from the reflectance values is done as follows:
g.region raster=lsat7_2002_toar.3 -p
i.vi blue=lsat7_2002_toar.1 red=lsat7_2002_toar.3 nir=lsat7_2002_toar.4 \
viname=arvi output=lsat7_2002.arvi
d.mon wx0
d.rast.leg lsat7_2002.arvi
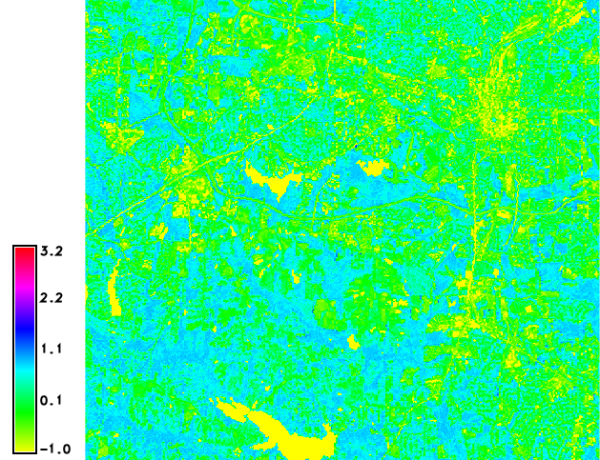
North Carolina dataset: ARVI
Calculation of GARI
The calculation of GARI from the reflectance values is done as follows:
g.region raster=lsat7_2002_toar.3 -p
i.vi blue=lsat7_2002_toar.1 green=lsat7_2002_toar.2 red=lsat7_2002_toar.3 \
nir=lsat7_2002_toar.4 viname=gari output=lsat7_2002.gari
d.mon wx0
d.rast.leg lsat7_2002.gari
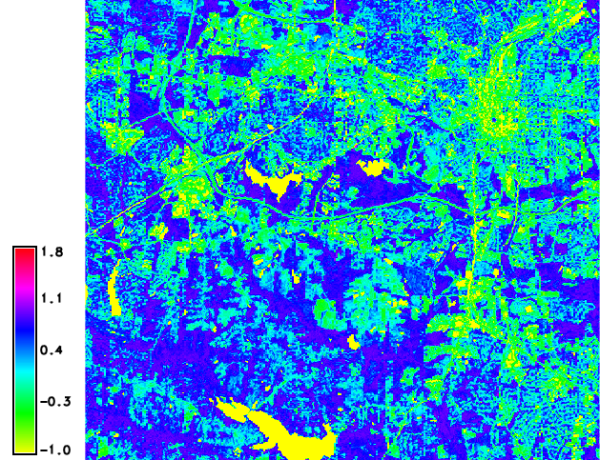
North Carolina dataset: GARI
NOTES
Originally from kepler.gps.caltech.edu (FAQ):
A FAQ on Vegetation in Remote Sensing
Written by Terrill W. Ray, Div. of Geological and Planetary Sciences,
California Institute of Technology, email: terrill@mars1.gps.caltech.edu
Snail Mail: Terrill Ray
Division of Geological and Planetary Sciences
Caltech, Mail Code 170-25
Pasadena, CA 91125
REFERENCES
AVHRR, Landsat TM5:- Bastiaanssen, W.G.M., 1995. Regionalization of surface flux densities and moisture indicators in composite terrain; a remote sensing approach under clear skies in mediterranean climates. PhD thesis, Wageningen Agricultural Univ., The Netherland, 271 pp. (PDF)
- Index DataBase: List of available Indices
SEE ALSO
i.albedo, i.aster.toar, i.landsat.toar, i.atcorr, i.tasscapAUTHORS
Baburao Kamble, Asian Institute of Technology, ThailandYann Chemin, Asian Institute of Technology, Thailand
SOURCE CODE
Available at: i.vi source code (history)
Latest change: Thursday Mar 23 20:37:09 2023 in commit: 064719b8da44cce254342ad02dad19b66160037b
Main index | Imagery index | Topics index | Keywords index | Graphical index | Full index
© 2003-2024 GRASS Development Team, GRASS GIS 8.3.3dev Reference Manual