
NAME
r.series - Makes each output cell value a function of the values assigned to the corresponding cells in the input raster map layers.KEYWORDS
raster, aggregation, series, parallelSYNOPSIS
Flags:
- -n
- Propagate NULLs
- -z
- Do not keep files open
- --overwrite
- Allow output files to overwrite existing files
- --help
- Print usage summary
- --verbose
- Verbose module output
- --quiet
- Quiet module output
- --ui
- Force launching GUI dialog
Parameters:
- input=name[,name,...]
- Name of input raster map(s)
- file=name
- Input file with one raster map name and optional one weight per line, field separator between name and weight is |
- output=name[,name,...] [required]
- Name for output raster map
- method=string[,string,...] [required]
- Aggregate operation
- Options: average, count, median, mode, minimum, min_raster, maximum, max_raster, stddev, range, sum, variance, diversity, slope, offset, detcoeff, tvalue, quart1, quart3, perc90, quantile, skewness, kurtosis
- quantile=float[,float,...]
- Quantile to calculate for method=quantile
- Options: 0.0-1.0
- weights=float[,float,...]
- Weighting factor for each input map, default value is 1.0 for each input map
- range=lo,hi
- Ignore values outside this range
- nprocs=integer
- Number of threads for parallel computing
- Default: 1
- memory=memory in MB
- Maximum memory to be used (in MB)
- Cache size for raster rows
- Default: 300
Table of contents
DESCRIPTION
r.series makes each output cell value a function of the values assigned to the corresponding cells in the input raster map layers.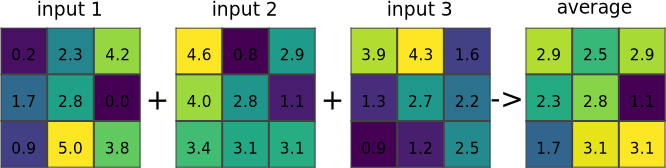
Figure: Illustration for an average of three input rasters
Following methods are available:
- average: average value
- count: count of non-NULL cells
- median: median value
- mode: most frequently occurring value
- minimum: lowest value
- min_raster: raster map number with the minimum time-series value
- maximum: highest value
- max_raster: raster map number with the maximum time-series value
- stddev: standard deviation
- range: range of values (max - min)
- sum: sum of values
- variance: statistical variance
- diversity: number of different values
- slope: linear regression slope
- offset: linear regression offset
- detcoeff: linear regression coefficient of determination
- tvalue: linear regression t-value
- quart1: first quartile
- quart3: third quartile
- perc90: ninetieth percentile
- quantile: arbitrary quantile
- skewness: skewness
- kurtosis: kurtosis
r.series input=map1,...,mapN \
output=map.mean,map.stddev \
method=average,stddev
r.series input=map1,...,mapN \
output=map.p10,map.p50,map.p90 \
method=quantile,quantile,quantile \
quantile=0.1,0.5,0.9
NOTES
No-data (NULL) handling
With -n flag, any cell for which any of the corresponding input cells are NULL is automatically set to NULL (NULL propagation). The aggregate function is not called, so all methods behave this way with respect to the -n flag.Without -n flag, the complete list of inputs for each cell (including NULLs) is passed to the aggregate function. Individual aggregates can handle data as they choose. Mostly, they just compute the aggregate over the non-NULL values, producing a NULL result only if all inputs are NULL.
Minimum and maximum analysis
The min_raster and max_raster methods generate a map with the number of the raster map that holds the minimum/maximum value of the time-series. The numbering starts at 0 up to n for the first and the last raster listed in input=, respectively.Range analysis
If the range= option is given, any values which fall outside that range will be treated as if they were NULL. The range parameter can be set to low,high thresholds: values outside of this range are treated as NULL (i.e., they will be ignored by most aggregates, or will cause the result to be NULL if -n is given). The low,high thresholds are floating point, so use -inf or inf for a single threshold (e.g., range=0,inf to ignore negative values, or range=-inf,-200.4 to ignore values above -200.4).Linear regression
Linear regression (slope, offset, coefficient of determination, t-value) assumes equal time intervals. If the data have irregular time intervals, NULL raster maps can be inserted into time series to make time intervals equal (see example).Quantiles
r.series can calculate arbitrary quantiles.Memory consumption
Memory usage is not an issue, as r.series only needs to hold one row from each map at a time.Management of open file limits
The maximum number of raster maps that can be processed is given by the user-specific limit of the operating system. For example, the soft limits for users are typically 1024 files. The soft limit can be changed with e.g.ulimit -n 4096 (UNIX-based operating systems) but it cannot be
higher than the hard limit. If the latter is too low, you can as superuser
add an entry in:
/etc/security/limits.conf # <domain> <type> <item> <value> your_username hard nofile 4096
cat /proc/sys/fs/file-max
For each map a weighting factor can be specified using the weights option. Using weights can be meaningful when computing the sum or average of maps with different temporal extent. The default weight is 1.0. The number of weights must be identical to the number of input maps and must have the same order. Weights can also be specified in the input file.
Use the -z flag to analyze large amounts of raster maps without hitting open files limit and the file option to avoid hitting the size limit of command line arguments. Note that the computation using the file option is slower than with the input option. For every single row in the output map(s) all input maps are opened and closed. The amount of RAM will rise linearly with the number of specified input maps. The input and file options are mutually exclusive: the former is a comma separated list of raster map names and the latter is a text file with a new line separated list of raster map names and optional weights. As separator between the map name and the weight the character "|" must be used.
Performance
To enable parallel processing, the user can specify the number of threads to be used with the nprocs parameter (default 1). The memory parameter (default 300 MB) can also be provided to determine the size of the buffer in MB for computation.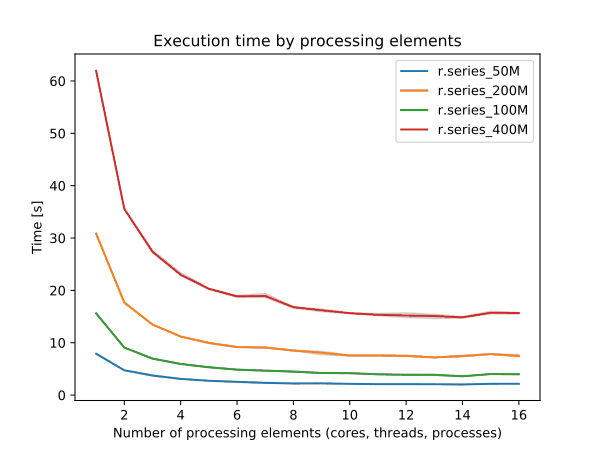
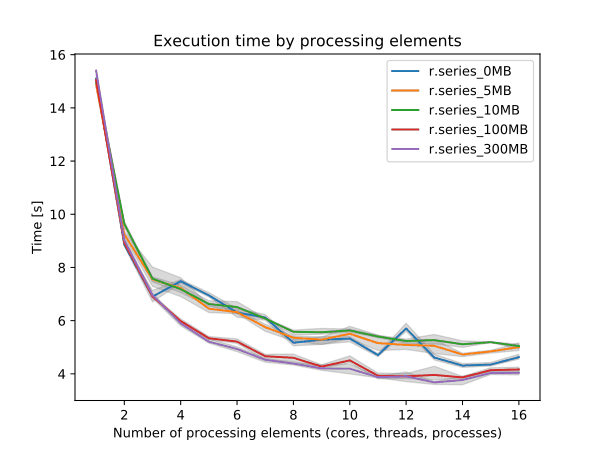
Figure: Benchmark on the left shows execution time for different number of cells, benchmark on the right shows execution time for different memory size for 10000x10000 raster. See benchmark scripts in source code. (Intel Core i9-10940X CPU @ 3.30GHz x 28)
To reduce the memory requirements to minimum, set option memory to zero. To take advantage of the parallelization, GRASS GIS needs to compiled with OpenMP enabled.
EXAMPLES
Using r.series with wildcards:
r.series input="`g.list pattern='insitu_data.*' sep=,`" \
output=insitu_data.stddev method=stddev
Note the g.list script also supports regular expressions for selecting map names.
Using r.series with NULL raster maps (in order to consider a
"complete" time series):
r.mapcalc "dummy = null()"
r.series in=map2001,map2002,dummy,dummy,map2005,map2006,dummy,map2008 \
out=res_slope,res_offset,res_coeff meth=slope,offset,detcoeff
Example for multiple aggregates to be computed in one run (3 resulting aggregates from two input maps):
r.series in=one,two out=result_avg,res_slope,result_count meth=sum,slope,count
Example to use the file option of r.series:
cat > input.txt << EOF map1 map2 map3 EOF r.series file=input.txt out=result_sum meth=sum
Example to use the file option of r.series including weights. The weight 0.75 should be assigned to map2. As the other maps do not have weights we can leave it out:
cat > input.txt << EOF map1 map2|0.75 map3 EOF r.series file=input.txt out=result_sum meth=sum
Example for counting the number of days above a certain temperature using daily average maps ('???' as DOY wildcard):
# Approach for shell based systems
r.series input=`g.list rast pattern="temp_2003_???_avg" separator=comma` \
output=temp_2003_days_over_25deg range=25.0,100.0 method=count
# Approach in two steps (e.g., for Windows systems)
g.list rast pattern="temp_2003_???_avg" output=mapnames.txt
r.series file=mapnames.txt \
output=temp_2003_days_over_25deg range=25.0,100.0 method=count
SEE ALSO
g.list, g.region, r.quantile, r.series.accumulate, r.series.interp, r.univarHints for large raster data processing
AUTHOR
Glynn ClementsSOURCE CODE
Available at: r.series source code (history)
Latest change: Tuesday May 06 23:45:42 2025 in commit: 59db85624e4fd204e389e27e1ddc28028dd3edfc
Main index | Raster index | Topics index | Keywords index | Graphical index | Full index
© 2003-2025 GRASS Development Team, GRASS GIS 8.4.3dev Reference Manual