NAME
r.maxent.train - Create and train a Maxent modelKEYWORDS
modeling, MaxentSYNOPSIS
Flags:
- -y
- Create a vector point layer from the sample predictions
- Import the file(s) with sample predictions as point feature layer.
- -b
- Create a vector point layer with predictions at background points
- Create a vector point layer with predictions at background points
- -g
- Create response curves.
- Create graphs showing how predicted relative probability of occurrence depends on the value of each environmental variable.
- -w
- Write response curve data to file
- Write output files containing the data used to make response curves, for import into external plotting software.
- -e
- Extrapolate
- Predict to regions of environmental space outside the limits encountered during training.
- -c
- Do not apply clamping
- Do not apply clamping when projecting.
- -f
- Fade effect clamping
- Reduce prediction at each point in projections by the difference between clamped and non-clamped output at that point.
- -l
- Disable linear features
- Do not use linear features for the model (they are used by default).
- -q
- Disable quadratic features
- Do not use quadratic features for the model (they are used by default).
- -p
- Disable product features
- Do not use product features for the model (they are used by default).
- -t
- Use threshold features
- By default, threshold features are not used. Use this flag to enable them.
- -h
- Disable hinge features
- Do not use hinge features for the model (they are used by default).
- -a
- Do not use automatic selection of feature classes
- By default, Maxent automatically selects which feature classes to use, based on number of training samples. Use this flag to disable autoselection of features.
- -n
- Don't add sample points to background if conditions differ
- By default, samples that have a combination of environmental values that isn't already present in the background are added to the background samples. Use this flag to avoid that.
- -j
- Use jackknife validation
- Measure importance of each environmental variable by training with each environmental variable first omitted, then used in isolation.
- -d
- Keep duplicate presence records.
- Keep duplicate presence records. If environmental data are in grids, duplicates are records in the same grid cell. Otherwise, duplicates are records with identical coordinates.
- -s
- Use a random seed
- If selected, a different random seed will be used for each run, so a different random test/train partition will be made and a different random subset of the background will be used, if applicable.
- -x
- Add all samples to the background
- Add all samples to the background, even if they have combinations of environmental values that are already present in the background
- -v
- Show the Maxent user interface
- Use this flag to show the Maxent interface. Note that when you select this option, Maxent will not start before you hit the start option.
- --overwrite
- Allow output files to overwrite existing files
- --help
- Print usage summary
- --verbose
- Verbose module output
- --quiet
- Quiet module output
- --ui
- Force launching GUI dialog
Parameters:
- samplesfile=name [required]
- Sample file presence locations
- Please enter the name of a file containing presence locations for one or more species.
- environmentallayersfile=name [required]
- Sample file with background locations
- Please enter the file name of the SWD file with environmental variables (can be created with v.maxent.swd or r.out.maxent_swd).
- togglelayertype=string
- Prefix that identifies categorical data
- Toggle continuous/categorical for environmental variables whose names begin with this prefix (default: all continuous)
- projectionlayers=name
- Location of folder with set of environmental variables.
- Location of an set of rasters representing the same environmental variables as used to create the Maxent model. They will be used to create a prediction layer based on the trained model.
- suffix=string
- Suffix for name(s) of prediction layer(s)
- Add a suffix to the name(s) of imported prediction layer(s)
- nodata=integer
- Nodata values
- Value to be interpreted as nodata values in SWD sample data
- Default: -9999
- outputdirectory=name [required]
- Directory where outputs will be written.
- Directory where outputs will be written. This should be different from the environmental layers directory.
- samplepredictions=name
- Name of sample prediction layer
- Give the name of sample prediction layer. If you leave this empty, the default name given by Maxent will be used.
- backgroundpredictions=name
- Name of background prediction layer
- Give the name of background prediction layer. If you leave this empty, the default name given by Maxent will be used.
- predictionlayer=name
- Name of raster prediction layer
- Give the name of raster prediction layer. If you leave this empty, the default name given by Maxent will be used.
- outputformat=string
- Representation probability
- Representation of probabilities used in writing output grids. See Help for details.
- Options: cloglog, logistic, cumulative, raw
- Default: cloglog
- betamultiplier=float
- Multiply all automatic regularization parameters by this number.
- Multiply all automatic regularization parameters by this number. A higher number gives a more spread-out distribution.
- Default: 1.0
- randomtestpoints=integer
- Percentage of random test points
- Percentage of presence localities to be randomly set aside as test points, used to compute the AUC, omission, etc.
- Default: 0
- testsamplesfile=name
- Test presence locations
- Use the presence localities in this csv file to compute statistics (AUC, omission, etc.).
- replicatetype=string
- Number of replicates in cross-validation
- If replicates > 1, do multiple runs using crossvalidate,bootstrap or subsample. See the Maxent help file for the difference.
- Options: crossvalidate, bootstrap, subsample
- Default: crossvalidate
- replicates=integer
- Number of replicates in cross-validation
- If replicates > 1, do multiple runs of this type: Crossvalidate: samples divided into replicates folds; each fold in turn used for test data. Bootstrap: replicate sample sets chosen by sampling with replacement. Subsample: replicate sample sets chosen by removing random test percentage without replacement to be used for evaluation.
- Options: 1-20
- Default: 1
- maximumiterations=integer
- Maximum iterations optimization
- Stop training after this many iterations of the optimization algorithm.
- Default: 500
- convergencethreshold=float
- Convergence threshold
- Stop training when the drop in log loss per iteration drops below this number.
- Default: 0.00005
- lq2lqptthreshold=integer
- Threshold for product and threshold features
- Number of samples at which product and threshold features start being used.
- Default: 80
- l2lqthreshold=integer
- Threshold for quadratic feature
- Number of samples at which quadratic features start being used.
- Default: 10
- hingethreshold=integer
- Threshold for hinge feature
- Number of samples at which hinge features start being used.
- Default: 15
- beta_threshold=float
- Regularization parameter for treshold features
- Regularization parameter to be applied to all threshold features; negative value enables automatic setting.
- Default: -1.0
- beta_categorical=float
- Regularization parameter for categorical features
- Regularization parameter to be applied to all categorical features; negative value enables automatic setting.
- Default: -1.0
- beta_lqp=float
- Regularization parameter for lin, quad and prod features
- Regularization parameter to be applied to all linear, quadratic and product features; negative value enables automatic setting.
- Default: -1.0
- beta_hinge=float
- Regularization parameter for hinge features
- Regularization parameter to be applied to all linear, quadratic and product features; negative value enables automatic setting.
- Default: -1.0
- defaultprevalence=float
- Default prevalence of the species
- Default prevalence of the species: probability of presence at ordinary occurrence points. See Elith et al., Diversity and Distributions, 2011 for details.
- Options: 0-1
- Default: 0.5
- maxent=name
- Location Maxent jar file
- Give the path to the Maxent executable file (maxent.jar)
- java=name
- Location java executable
- If Java is installed, but cannot be found, the user can provide the path to the java executable file. Note, an alternative is to use the r.maxent.setup addon.
- threads=integer
- Number of processor threads to use.
- Number of threads for parallel computing
- Default: 1
- memory=memory in MB
- Maximum memory to be used (in MB)
- Maximum memory to be used by Maxent (in MB)
- Default: 300
- precision=integer
- Precision suitability map
- Set the required precision (in the form of number of decimal digits) of the species suitability raster layer (leave empty for default).
Table of contents
DESCRIPTION
The r.maxent.train module is a front-end to the Maxent software, providing a convenient way to run the Maxent software, and create output layers in GRASS GIS.

A workflow, from data preparation, training a model to model prediction using three GRASS GIS addons.
It is part of a set of three addons that can be used to prepare the input data for the Maxent model (v.maxent.sdm), to train a maxent presence only model (this module), and to use the model to create prediction layers (r.maxent.predict). With r.maxent.train a Maxent presence only model can be created using the Maxent software. As input, the addon requires two comma-separated files, one with the species locations and another of background points locations. Both need to include columns with the X, Y and sample values of the environmental variables that you want to use as predictor variables. You can use the r.out.maxent_swd or v.maxent.swd addons to create these files. For more details about the structure of these files, see the Maxent website.
The only other requirement is to provide an output folder. With these inputs, a Maxent model will be created. If you also provide a folder with environmental raster layers with names corresponding to the names of the environmental variables in the SWD files, the module will create a prediction (suitability distribution) raster layer as well.
Note that the Maxent software generates ASCII files without projection information. That means you need to make sure yourself that the environmental layers you provide are in the same reference coordinate system as your current mapset. An easy way to ensure this is by using the v.maxent_swd from the same mapset to create those input environmental layers for Maxent. See the workflow in the Examples.
The addon provides access to nearly all parameters available in the Maxent software. On the above-mentioned website, you can find a tutorial that explains most of these options. For the other options, see the Maxent help file.
NOTES
This addon requires the Maxent software (version ≥ 3.4). You can download the software from the Maxent website. The r.maxent.setup module provides an helper function to enable GRASS GIS to use the Maxent software.The r.maxent.train addon runs Maxent in the background. If you want to check the Maxent settings first, you can set the -v flag to open the Maxent user interface with all parameters filled in. You will need to hit the Run button to actually run Maxent.
Besides the files directly generated by Maxent, the addon creates the maxent_explanatory_variable_names.csv file. This file contains the names of the model explanatory variables. You can use this when you quickly want to check the names of the explanatory variables, e.g., when using r.maxent.predict.
EXAMPLE
The examples below use a dataset that you can download from here. It includes a vector point layer with observation locations of the pale-throated sloth (Bradypus tridactylus) from GBIF, the IUCN RedList range map of the species, a boundary layer of the South American countries from NaturalEarth and a number of bioclim raster layers from WorldClim version 2.1, representing the climate conditions representing the period 1970-2000 and the climate conditions predicted for 2061–2080 based on the GCM BCC-CSM2-MR and SSP 585.
The zip file contains a folder sampledata. This is a location with five subfolders PERMANENT, sloth, current, future and model01. Copy this Location to a GRASS Database (use an existing one or create one first). If you are not familiar with the concept of Locations and Mapsets, please first read the explanation about the GRASS GIS database.
Unzip the file, start up GRASS GIS, open the GRASS GIS database to which you copied the folder sampledata, switch to the Location sampledata and open the mapset model01. This mapset should have access to the other mapsets.
1: Data preparation
You can use the v.maxent.swd to create the required input layers. The code below creates the SWD file with the locations where the species has been recorded (species_output) and a SWD file with randomly created background point locations (bgr_ouput). The SWD files contain, for each location, the values of the raster layers selected with the evp_maps parameter. With the parameter export_rasters you tell the addon to export the raster layers as well.v.maxent.swd -t \ species=Bradypus_tridactylus \ evp_maps=bio02,bio03,bio08,bio09,bio13,bio15,bio17 \ evp_cat=sa_eco_l2@current \ alias_cat=landuse \ nbgp=10000 \ bgr_output=bgrd_swd.csv \ species_output=spec_swd.csv \ export_rasters=envlayers
The output is a folder with the so-called SWD files with the XY coordinates for the species presence location (spec_swd.csv) and the background locations (bgrd_swd.csv. Both also include the values of the input raster layers for the given point locations. In addition, there is the subfolder envlayers with the environmental raster layers in ascii format.
2: Train the model
Use the output of v.maxent.swd as input for r.maxent.train. First create a subfolder output_model1, so we can write the output to that folder.The projectionlayers parameter is optionally. If you set it, a raster prediction layer will be created that represent the potential suitability distribution under current conditions (the conditions used to train the model).
With the -y and -b flags the point layers with the sample predictions and the predictions at the background point locations are created. Their values correspond to the values of the raster prediction layer.
r.maxent.train -y -b -g \ samplesfile=spec_swd.csv \ environmentallayersfile=bgrd_swd.csv \ togglelayertype=landuse \ projectionlayers=envlayers \ samplepredictions=model_1_samplepred \ backgroundpredictions=model_1_bgrdpred \ predictionlayer=model_1_suitability_current \ outputdirectory=output_model1
When r.maxent.train is finished, go to the output folder and open the Bradypus_tridactylus.html file for an explanation of the different model outputs and model evaluation statistics. For a more detailed explanation, see the tutorial on the Maxent website.
In your current mapset, you'll find the raster prediction layer, and the sample and background point layers with the predicted values.
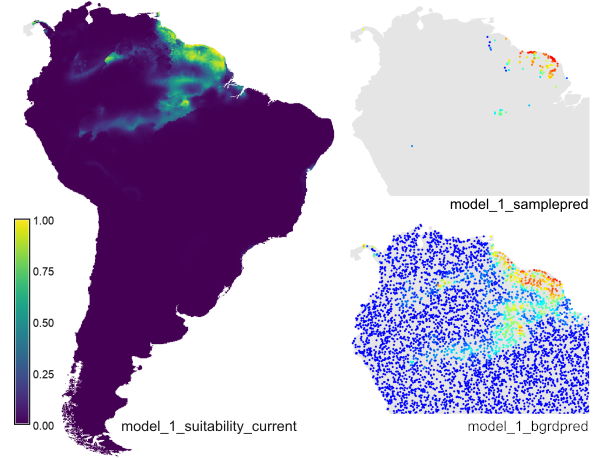
The example creates the prediction raster layer 'model_1_suitability_current', the sample point layer 'model_1_samplepred' and the background point layer 'model_bgrdpred' (for the latter, only part of the map is shown here).
3: Create a prediction layer
The third step is to use the model created in the previous step to predict the species suitability distribution under future climates. Note, we are going to make the (unrealistic) assumption that the ecosystems do not change.r.maxent.predict lambda=output_model1/Bradypus_tridactylus.lambdas \ rasters=bio02_ssp585,bio03_ssp585,bio08_ssp585,bio09_ssp585,bio13_ssp585,bio15_ssp585,bio17_ssp585,sa_eco_l2 \ variables=bio02,bio03,bio08,bio09,bio13,bio15,bio17,landuse \ output=model_1_ssp585
The resulting layer is written to the current mapset as model_1_ssp585 (right map in the figure below). The results suggest the area with suitable conditions will increase under future climates compared the that under the current conditions (left map in the figure below). This result is unexpected, and warrants further investigation.
REFERENCES
- Steven J. Phillips, Miroslav Dudík, Robert E. Schapire. 2020: Maxent software for modeling species niches and distributions (Version 3.4.1). Available from url: https://biodiversityinformatics.amnh.org/open_source/maxent and https://github.com/mrmaxent/Maxent
- Steven J. Phillips, Miroslav Dudík, Robert E. Schapire. 2004: A maximum entropy approach to species distribution modeling. In Proceedings of the Twenty-First International Conference on Machine Learning, pages 655-662, 2004.
- Steven J. Phillips, Robert P. Anderson, Robert E. Schapire. 2006: Maximum entropy modeling of species geographic distributions. Ecological Modelling, 190:231-259, 2006.
- Jane Elith, Steven J. Phillips, Trevor Hastie, Miroslav Dudík, Yung En Chee, Colin J. Yates. 2011: A statistical explanation of MaxEnt for ecologists. Diversity and Distributions, 17:43-57, 2011.
SEE ALSO
- v.maxent.swd, creating species and background swd files and prediction rasters that can be used directly by the r.maxent.train addon (or the Maxent software itself) to create species distribution models.
- r.out.maxent_swd, creating species and background swd files based on species distribution data in raster format.
- r.maxent.predict, creating a suitability layer based on a set of environmental layers and a Maxent model, e.g., created using the r.maxent.train addon.
- r.maxent.setup, helper function to allow GRASS to use Maxent.
AUTHOR
Paulo van Breugel, HAS green academy, Innovative Biomonitoring research group, Climate-robust Landscapes research groupSOURCE CODE
Available at: r.maxent.train source code (history)
Latest change: Thursday Jan 15 21:45:55 2026 in commit: 2a1ecddccd7274a102dc65d6588834d05558ab84
Main index | Raster index | Topics index | Keywords index | Graphical index | Full index
© 2003-2025 GRASS Development Team, GRASS GIS 8.4.3dev Reference Manual