r.resamp.filter
Resamples raster map layers using an analytic kernel.
r.resamp.filter [-n] input=name output=name filter=string [,string,...] [radius=float [,float,...]] [x_radius=float [,float,...]] [y_radius=float [,float,...]] [memory=memory in MB] [nprocs=integer] [--overwrite] [--verbose] [--quiet] [--qq] [--ui]
Example:
r.resamp.filter input=name output=name filter=box
grass.script.run_command("r.resamp.filter", input, output, filter, radius=None, x_radius=None, y_radius=None, memory=300, nprocs=0, flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
gs.run_command("r.resamp.filter", input="name", output="name", filter="box")
grass.tools.Tools.r_resamp_filter(input, output, filter, radius=None, x_radius=None, y_radius=None, memory=300, nprocs=0, flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
tools = Tools()
tools.r_resamp_filter(input="name", output="name", filter="box")
This grass.tools API is experimental in version 8.5 and expected to be stable in version 8.6.
Parameters
input=name [required]
Name of input raster map
output=name [required]
Name for output raster map
filter=string [,string,...] [required]
Filter kernel(s)
Allowed values: box, bartlett, gauss, normal, hermite, sinc, lanczos1, lanczos2, lanczos3, hann, hamming, blackman
radius=float [,float,...]
Filter radius
x_radius=float [,float,...]
Filter radius (horizontal)
y_radius=float [,float,...]
Filter radius (vertical)
memory=memory in MB
Maximum memory to be used (in MB)
Cache size for raster rows
Default: 300
nprocs=integer
Number of threads for parallel computing
0: use OpenMP default; >0: use nprocs; <0: use MAX-nprocs
Default: 0
-n
Propagate NULLs
--overwrite
Allow output files to overwrite existing files
--help
Print usage summary
--verbose
Verbose module output
--quiet
Quiet module output
--qq
Very quiet module output
--ui
Force launching GUI dialog
input : str, required
Name of input raster map
Used as: input, raster, name
output : str, required
Name for output raster map
Used as: output, raster, name
filter : str | list[str], required
Filter kernel(s)
Allowed values: box, bartlett, gauss, normal, hermite, sinc, lanczos1, lanczos2, lanczos3, hann, hamming, blackman
radius : float | list[float] | str, optional
Filter radius
x_radius : float | list[float] | str, optional
Filter radius (horizontal)
y_radius : float | list[float] | str, optional
Filter radius (vertical)
memory : int, optional
Maximum memory to be used (in MB)
Cache size for raster rows
Used as: memory in MB
Default: 300
nprocs : int, optional
Number of threads for parallel computing
0: use OpenMP default; >0: use nprocs; <0: use MAX-nprocs
Default: 0
flags : str, optional
Allowed values: n
n
Propagate NULLs
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
input : str | np.ndarray, required
Name of input raster map
Used as: input, raster, name
output : str | type(np.ndarray) | type(np.array) | type(gs.array.array), required
Name for output raster map
Used as: output, raster, name
filter : str | list[str], required
Filter kernel(s)
Allowed values: box, bartlett, gauss, normal, hermite, sinc, lanczos1, lanczos2, lanczos3, hann, hamming, blackman
radius : float | list[float] | str, optional
Filter radius
x_radius : float | list[float] | str, optional
Filter radius (horizontal)
y_radius : float | list[float] | str, optional
Filter radius (vertical)
memory : int, optional
Maximum memory to be used (in MB)
Cache size for raster rows
Used as: memory in MB
Default: 300
nprocs : int, optional
Number of threads for parallel computing
0: use OpenMP default; >0: use nprocs; <0: use MAX-nprocs
Default: 0
flags : str, optional
Allowed values: n
n
Propagate NULLs
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
Returns:
result : grass.tools.support.ToolResult | np.ndarray | tuple[np.ndarray] | None
If the tool produces text as standard output, a ToolResult object will be returned. Otherwise, None will be returned. If an array type (e.g., np.ndarray) is used for one of the raster outputs, the result will be an array and will have the shape corresponding to the computational region. If an array type is used for more than one raster output, the result will be a tuple of arrays.
Raises:
grass.tools.ToolError: When the tool ended with an error.
DESCRIPTION
r.resamp.filter resamples an input raster, filtering the input with an analytic kernel. Each output cell is typically calculated based upon a small subset of the input cells, not the entire input. r.resamp.filter performs convolution (i.e. a weighted sum is calculated for every raster cell).
The radii must be given in map units. In order to consider more than one input cell, at least one finite radius must be larger than half the resolution of the input map, otherwise the selected kernels will have no effect.
The module maps the input range to the width of the window function, so wider windows will be "sharper" (have a higher cut-off frequency), e.g. lanczos3 will be sharper than lanczos2.
r.resamp.filter implements FIR (finite impulse response) filtering. All of the functions are low-pass filters, as they are symmetric. See Wikipedia: Window function for examples of common window functions and their frequency responses.
A piecewise-continuous function defined by sampled data can be considered a mixture (sum) of the underlying signal and quantisation noise. The intent of a low pass filter is to discard the quantisation noise while retaining the signal. The cut-off frequency is normally chosen according to the sampling frequency, as the quantisation noise is dominated by the sampling frequency and its harmonics. In general, the cut-off frequency is inversely proportional to the width of the central "lobe" of the window function.
When using r.resamp.filter with a specific radius, a specific cut-off frequency regardless of the method is chosen. So while lanczos3 uses 3 times as large a window as lanczos1, the cut-off frequency remains the same. Effectively, the radius is "normalised".
All of the kernels specified by the filter parameter are multiplied together. Typical usage will use either a single finitie window or an infinite kernel along with a finite window.
Usage hints
To smooth a map, keeping its resolution, a good starting point is to use the filters gauss,box with the radii 0.5 * input resolution, 2 * input resolution. See also r.neighbors
When resampling a map to a higher resolution (alternative to interpolation, e.g. r.resamp.interp), a good starting point is to use the filters gauss,box with the radii 1.5 * input resolution, 3 * input resolution.
When resampling a map to a lower resolution (alternative to aggregation, e.g. r.resamp.stats), a good starting point is to use the filters gauss,box with the radii 0.25 * output resolution, 1 * output resolution.
These are recommendations for initial settings. The selection of filters and radii might need adjustment according to the actual purpose.
NOTES
Resampling modules (r.resample, r.resamp.stats, r.resamp.interp, r.resamp.rst, r.resamp.filter) resample the map to match the current region settings.
When using a kernel which can have negative values (sinc, Lanczos), the -n flag should be used. Otherwise, extreme values can arise due to the total weight being close (or even equal) to zero.
Kernels with infinite extent (Gauss, normal, sinc, Hann, Hamming, Blackman) must be used in conjunction with a finite windowing function (box, Bartlett, Hermite, Lanczos).
The way that Lanczos filters are defined, the number of samples is supposed to be proportional to the order ("a" parameter), so lanczos3 should use 3 times as many samples (at the same sampling frequency, i.e. cover 3 times as large a time interval) as lanczos1 in order to get a similar frequency response (higher-order filters will fall off faster, but the frequency at which the fall-off starts should be the same). See Wikipedia: Lanczos-kernel.svg for an illustration. If both graphs were drawn on the same axes, they would have roughly the same shape, but the a=3 window would have a longer tail. By scaling the axes to the same width, the a=3 window has a narrower central lobe.
For longitude-latitude coordinate reference systems, the interpolation algorithm is based on degree fractions, not on the absolute distances between cell centers. Any attempt to implement the latter would violate the integrity of the interpolation method.
PERFORMANCE
By specifying the number of parallel processes with nprocs option, r.resamp.filter can run faster, see benchmarks below.
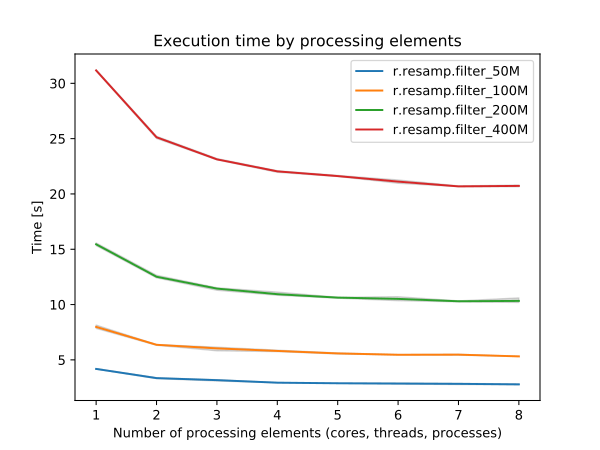
Figure: Benchmark shows execution time for different number of cells.
See benchmark script in the source code.
To reduce the memory requirements to minimum, set option memory to zero. To take advantage of the parallelization, GRASS needs to compiled with OpenMP enabled.
SEE ALSO
g.region, r.mfilter, r.resample, r.resamp.interp, r.resamp.rst, r.resamp.stats
Overview: Interpolation and Resampling in GRASS
AUTHOR
Glynn Clements
SOURCE CODE
Available at: r.resamp.filter source code
(history)
Latest change: Friday Aug 08 09:39:19 2025 in commit ed72c71