r.profile
Outputs the raster map layer values lying on user-defined line(s).
r.profile [-gc] input=name [output=name] [coordinates=east,north [,east,north,...]] [file=name] [resolution=float] [null_value=string] [units=string] format=name [color_format=name] [separator=character] [--overwrite] [--verbose] [--quiet] [--qq] [--ui]
Example:
r.profile input=name format=plain
grass.script.parse_command("r.profile", input, output="-", coordinates=None, file=None, resolution=None, null_value="*", units=None, format="plain", color_format=None, separator="comma", flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
gs.parse_command("r.profile", input="name", format="json")
grass.tools.Tools.r_profile(input, output="-", coordinates=None, file=None, resolution=None, null_value="*", units=None, format="plain", color_format=None, separator="comma", flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
tools = Tools()
tools.r_profile(input="name", format="json")
This grass.tools API is experimental in version 8.5 and expected to be stable in version 8.6.
Parameters
input=name [required]
Name of input raster map
output=name
Name of file for output (use output=- for stdout)
Default: -
coordinates=east,north [,east,north,...]
Profile coordinate pairs
file=name
Name of input file containing coordinate pairs
Use instead of the 'coordinates' option. "-" reads from stdin.
resolution=float
Resolution along profile (default = current region resolution)
null_value=string
String representing NULL value
Default: *
units=string
Units
If units are not specified, current project units are used. Meters are used by default in geographic (latlon) projects.
Allowed values: meters, kilometers, feet, miles
format=name [required]
Output format
Allowed values: plain, csv, json
Default: plain
plain: Human readable text output
csv: CSV (Comma Separated Values)
json: JSON (JavaScript Object Notation)
color_format=name
Color format
Color format for output values.
Allowed values: rgb, hex, hsv, triplet
rgb: output color in RGB format
hex: output color in HEX format
hsv: output color in HSV format (experimental)
triplet: output color in colon-separated RGB format
separator=character
Field separator
Special characters: pipe, comma, space, tab, newline
Default: comma
-g
Output easting and northing in first two columns of four column output
-c
Output color values for each profile point (format controlled by color_format option; default is 'triplet' for plain output, 'hex' for JSON)
--overwrite
Allow output files to overwrite existing files
--help
Print usage summary
--verbose
Verbose module output
--quiet
Quiet module output
--qq
Very quiet module output
--ui
Force launching GUI dialog
input : str, required
Name of input raster map
Used as: input, raster, name
output : str, optional
Name of file for output (use output=- for stdout)
Used as: output, file, name
Default: -
coordinates : list[tuple[float, float]] | tuple[float, float] | list[float] | str, optional
Profile coordinate pairs
Used as: input, coords, east,north
file : str, optional
Name of input file containing coordinate pairs
Use instead of the 'coordinates' option. "-" reads from stdin.
Used as: input, file, name
resolution : float, optional
Resolution along profile (default = current region resolution)
null_value : str, optional
String representing NULL value
Used as: string
Default: *
units : str, optional
Units
If units are not specified, current project units are used. Meters are used by default in geographic (latlon) projects.
Allowed values: meters, kilometers, feet, miles
format : str, required
Output format
Used as: name
Allowed values: plain, csv, json
plain: Human readable text output
csv: CSV (Comma Separated Values)
json: JSON (JavaScript Object Notation)
Default: plain
color_format : str, optional
Color format
Color format for output values.
Used as: name
Allowed values: rgb, hex, hsv, triplet
rgb: output color in RGB format
hex: output color in HEX format
hsv: output color in HSV format (experimental)
triplet: output color in colon-separated RGB format
separator : str, optional
Field separator
Special characters: pipe, comma, space, tab, newline
Used as: input, separator, character
Default: comma
flags : str, optional
Allowed values: g, c
g
Output easting and northing in first two columns of four column output
c
Output color values for each profile point (format controlled by color_format option; default is 'triplet' for plain output, 'hex' for JSON)
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
input : str | np.ndarray, required
Name of input raster map
Used as: input, raster, name
output : str, optional
Name of file for output (use output=- for stdout)
Used as: output, file, name
Default: -
coordinates : list[tuple[float, float]] | tuple[float, float] | list[float] | str, optional
Profile coordinate pairs
Used as: input, coords, east,north
file : str | io.StringIO, optional
Name of input file containing coordinate pairs
Use instead of the 'coordinates' option. "-" reads from stdin.
Used as: input, file, name
resolution : float, optional
Resolution along profile (default = current region resolution)
null_value : str, optional
String representing NULL value
Used as: string
Default: *
units : str, optional
Units
If units are not specified, current project units are used. Meters are used by default in geographic (latlon) projects.
Allowed values: meters, kilometers, feet, miles
format : str, required
Output format
Used as: name
Allowed values: plain, csv, json
plain: Human readable text output
csv: CSV (Comma Separated Values)
json: JSON (JavaScript Object Notation)
Default: plain
color_format : str, optional
Color format
Color format for output values.
Used as: name
Allowed values: rgb, hex, hsv, triplet
rgb: output color in RGB format
hex: output color in HEX format
hsv: output color in HSV format (experimental)
triplet: output color in colon-separated RGB format
separator : str, optional
Field separator
Special characters: pipe, comma, space, tab, newline
Used as: input, separator, character
Default: comma
flags : str, optional
Allowed values: g, c
g
Output easting and northing in first two columns of four column output
c
Output color values for each profile point (format controlled by color_format option; default is 'triplet' for plain output, 'hex' for JSON)
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
Returns:
result : grass.tools.support.ToolResult | None
If the tool produces text as standard output, a ToolResult object will be returned. Otherwise, None will be returned.
Raises:
grass.tools.ToolError: When the tool ended with an error.
DESCRIPTION
This program outputs two or four column (with -g) data to stdout or an ASCII file. The default two column output consists of cumulative profile length and raster value. The optional four column output consists of easting, northing, cumulative profile length, and raster value. Profile end or "turning" points can be set manually with the coordinates argument. The profile resolution, or distance between profile points, is obtained from the current region resolution, or can be manually set with the resolution argument.
The coordinates parameter can be set to comma separated geographic
coordinates for profile line endpoints. Alternatively the coordinate
pairs can be piped from the text file specified by file option, or
if set to "-", from stdin. In these cases the coordinate pairs should
be given one comma separated pair per line.
The resolution parameter sets the distance between each profile point (resolution). The resolution must be provided in GRASS database units (i.e. decimal degrees for Lat Long databases and meters for UTM). By default r.profile uses the resolution of the current GRASS region.
The null parameter can optionally be set to change the character string representing null values.
The optional color output (with -c) provides the associated GRASS colour value for each profile point. The format of the color output is controlled by the color_format option, which can be set to hex, triplet, rgb, or hsv color formats. The default color format is triplet for plain output, and hex for JSON output.
NOTES
The profile resolution is measured exactly from the supplied end or "turning" point along the profile. The end of a profile segment will be an exact multiple of the profile resolution and will therefore not always match the end point coordinates entered for the segmanet.
To extract the numbers in scripts, following parameters can be used:
r.profile input=dgm12.5 coordinates=3570631,5763556 2>/dev/null
This filters out everything except the numbers.
Option units enables to set units of the profile length output. If the units are not specified, current coordinate reference system's units will be used. In case of geographic CRS (latitude/longitude), meters are used as default unit.
EXAMPLES
Extraction of values along profile defined by coordinates (variant 1)
Extract a profile with coordinates (waypoints) provided on the command line (North Carolina data set):
g.region raster=elevation -p
r.profile -g input=elevation output=profile_points.csv \
coordinates=641712,226095,641546,224138,641546,222048,641049,221186
This will extract a profile along the track defined by the three coordinate pairs. The output file "profile_points.csv" contains east,north,distance,value (here: elevation).
Extraction of values along profile defined by coordinates (variant 2)
Coordinate pairs can also be "piped" into r.profile (variant 2a):
r.profile elevation resolution=1000 file=- << EOF
641712,226095
641546,224138
641546,222048
641049,221186
EOF
Coordinate pairs can also be "piped" into r.profile (variant 2b):
echo "641712,226095
641546,224138
641546,222048
641049,221186" > coors.txt
cat coors.txt | r.profile elevation resolution=1000 file=-
The output is printed into the terminal (unless the output parameter is used) and looks as follows:
Using resolution: 1000 [meters]
Output columns:
Along track dist. [meters], Elevation
Approx. transect length: 1964.027749 [meters]
0.000000 84.661507
1000.000000 98.179062
Approx. transect length: 2090.000000 [meters]
1964.027749 83.638138
2964.027749 89.141029
3964.027749 78.497757
Approx. transect length: 995.014070 [meters]
4054.027749 73.988029
JSON Output
r.profile -g input=elevation coordinates=641712,226095,641546,224138,641546,222048,641049,221186 -c format=json resolution=1000
The output looks as follows:
[
{
"easting": 641712,
"northing": 226095,
"distance": 0,
"value": 84.661506652832031,
"color": "#71FF00"
},
{
"easting": 641627.47980925441,
"northing": 225098.57823319823,
"distance": 1000.0000000000125,
"value": 98.179061889648438,
"color": "#FFF100"
},
{
"easting": 641546,
"northing": 224138,
"distance": 1964.0277492948007,
"value": 83.638137817382812,
"color": "#64FF00"
},
{
"easting": 641546,
"northing": 223138,
"distance": 2964.0277492948007,
"value": 89.141029357910156,
"color": "#A9FF00"
},
{
"easting": 641546,
"northing": 222138,
"distance": 3964.0277492948007,
"value": 78.497756958007812,
"color": "#23FF00"
},
{
"easting": 641546,
"northing": 222048,
"distance": 4054.0277492948007,
"value": 73.988029479980469,
"color": "#00F911"
}
]
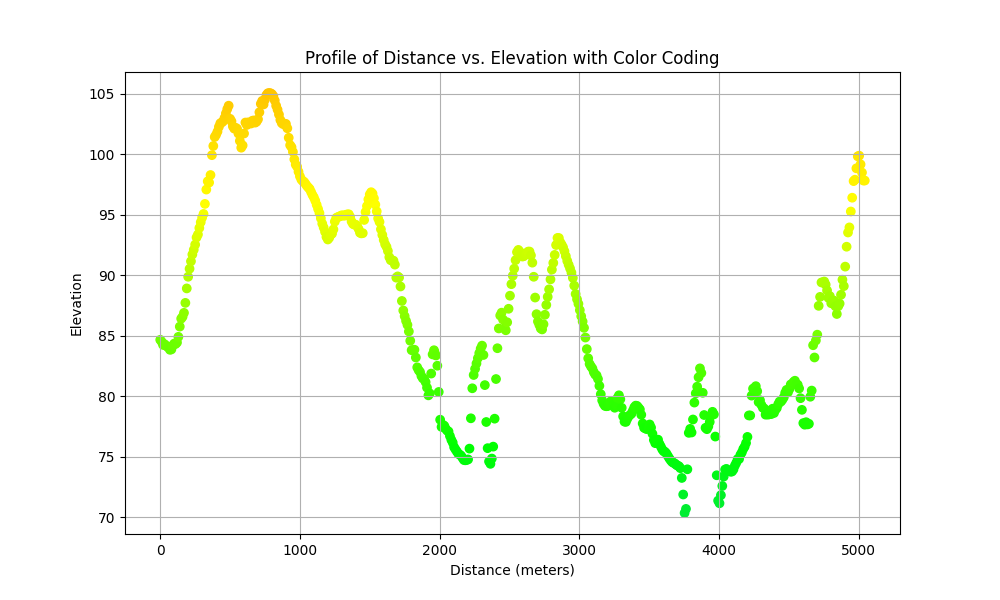
Using JSON output with Python for plotting data
The JSON output makes for ease of integration with popular python data science libraries. For instance, here is an example of creating a scatterplot of distance vs elevation with color coding.
import grass.script as gs
import pandas as pd
import matplotlib.pyplot as plt
# Run r.profile command
elevation = gs.read_command(
"r.profile",
input="elevation",
coordinates="641712,226095,641546,224138,641546,222048,641049,221186",
format="json",
flags="gc",
)
# Load the JSON data into a dataframe
df = pd.read_json(elevation)
# Create the scatter plot
plt.figure(figsize=(10, 6))
plt.scatter(df["distance"], df["value"], c=df["color"], marker="o")
plt.title("Profile of Distance vs. Elevation with Color Coding")
plt.xlabel("Distance (meters)")
plt.ylabel("Elevation")
plt.grid(True)
plt.show()
Using output in other programs
The multi column output from r.profile is intended for easy use in other programs. The output can be piped (|) directly into other programs or saved to a file for later use. Output with geographic coordinates (-g) is compatible with v.in.ascii and can be piped directly into this program.
r.profile -g input=elevation coordinates=... | v.in.ascii output=elevation_profile separator=space
SEE ALSO
v.in.ascii, r.what, r.transect, wxGUI profile tool
AUTHOR
SOURCE CODE
Available at: r.profile source code
(history)
Latest change: Thursday Oct 02 14:39:13 2025 in commit 1850489