v.cluster
Performs cluster identification.
v.cluster [-2bt] input=name output=name [layer=string] [distance=float] [min=integer] [method=string] [--overwrite] [--verbose] [--quiet] [--qq] [--ui]
Example:
v.cluster input=name output=name
grass.script.run_command("v.cluster", input, output, layer="2", distance=None, min=None, method="dbscan", flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
gs.run_command("v.cluster", input="name", output="name")
grass.tools.Tools.v_cluster(input, output, layer="2", distance=None, min=None, method="dbscan", flags=None, overwrite=None, verbose=None, quiet=None, superquiet=None)
Example:
tools = Tools()
tools.v_cluster(input="name", output="name")
This grass.tools API is experimental in version 8.5 and expected to be stable in version 8.6.
Parameters
input=name [required]
Name of input vector map
Or data source for direct OGR access
output=name [required]
Name for output vector map
layer=string
Layer number or name for cluster ids
Vector features can have category values in different layers. This number determines which layer to use. When used with direct OGR access this is the layer name.
Default: 2
distance=float
Maximum distance to neighbors
min=integer
Minimum number of points to create a cluster
method=string
Clustering method
Allowed values: dbscan, dbscan2, density, optics, optics2
Default: dbscan
-2
Force 2D clustering
-b
Do not build topology
Advantageous when handling a large number of points
-t
Do not create attribute table
--overwrite
Allow output files to overwrite existing files
--help
Print usage summary
--verbose
Verbose module output
--quiet
Quiet module output
--qq
Very quiet module output
--ui
Force launching GUI dialog
input : str, required
Name of input vector map
Or data source for direct OGR access
Used as: input, vector, name
output : str, required
Name for output vector map
Used as: output, vector, name
layer : str, optional
Layer number or name for cluster ids
Vector features can have category values in different layers. This number determines which layer to use. When used with direct OGR access this is the layer name.
Used as: input, layer
Default: 2
distance : float, optional
Maximum distance to neighbors
min : int, optional
Minimum number of points to create a cluster
method : str, optional
Clustering method
Allowed values: dbscan, dbscan2, density, optics, optics2
Default: dbscan
flags : str, optional
Allowed values: 2, b, t
2
Force 2D clustering
b
Do not build topology
Advantageous when handling a large number of points
t
Do not create attribute table
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
input : str, required
Name of input vector map
Or data source for direct OGR access
Used as: input, vector, name
output : str, required
Name for output vector map
Used as: output, vector, name
layer : str, optional
Layer number or name for cluster ids
Vector features can have category values in different layers. This number determines which layer to use. When used with direct OGR access this is the layer name.
Used as: input, layer
Default: 2
distance : float, optional
Maximum distance to neighbors
min : int, optional
Minimum number of points to create a cluster
method : str, optional
Clustering method
Allowed values: dbscan, dbscan2, density, optics, optics2
Default: dbscan
flags : str, optional
Allowed values: 2, b, t
2
Force 2D clustering
b
Do not build topology
Advantageous when handling a large number of points
t
Do not create attribute table
overwrite : bool, optional
Allow output files to overwrite existing files
Default: None
verbose : bool, optional
Verbose module output
Default: None
quiet : bool, optional
Quiet module output
Default: None
superquiet : bool, optional
Very quiet module output
Default: None
Returns:
result : grass.tools.support.ToolResult | None
If the tool produces text as standard output, a ToolResult object will be returned. Otherwise, None will be returned.
Raises:
grass.tools.ToolError: When the tool ended with an error.
DESCRIPTION
v.cluster partitions a point cloud into clusters or clumps.
If the minimum number of points is not specified with the min option, the minimum number of points to constitute a cluster is number of dimensions + 1, i.e. 3 for 2D points and 4 for 3D points.
If the maximum distance is not specified with the distance option, the maximum distance is estimated from the observed distances to the neighbors using the upper 99% confidence interval.
v.cluster supports different methods for clustering. The recommended methods are method=dbscan if all clusters should have a density (maximum distance between points) not larger than distance or method=density if clusters should be created separately for each observed density (distance to the farthest neighbor).
Clustering methods
dbscan method
The Density-Based Spatial Clustering of Applications with Noise is a commonly used clustering algorithm. A new cluster is started for a point with at least min - 1 neighbors within the maximum distance. These neighbors are added to the cluster. The cluster is then expanded as long as at least min - 1 neighbors are within the maximum distance for each point already in the cluster.
dbscan2 method
Similar to dbscan, but here it is sufficient if the resultant cluster consists of at least min points, even if no point in the cluster has at least min - 1 neighbors within distance.
density method
This method creates clusters according to their point density. The maximum distance is not used. Instead, the points are sorted ascending by the distance to their farthest neighbor (core distance), inspecting min - 1 neighbors. The densest cluster is created first, using as threshold the core distance of the seed point. The cluster is expanded as for DBSCAN, with the difference that each cluster has its own maximum distance. This method can identify clusters with different densities and can create nested clusters.
optics method
This method is Ordering Points to Identify the Clustering Structure. It is controlled by the number of neighbor points (option min - 1). The core distance of a point is the distance to the farthest neighbor. The reachability of a point q is its distance from a point p (original optics: max(core-distance(p), distance(p, q))). The aim of the optics method is to reduce the reachability of each point. Each unprocessed point is the seed for a new cluster. Its neighbors are added to a queue sorted by smallest reachability if their reachability can be reduced. The points in the queue are processed and their unprocessed neighbors are added to a queue sorted by smallest reachability if their reachability can be reduced.
The optics method does not create clusters itself, but produces an ordered list of the points together with their reachability. The output list is ordered according to the order of processing: the first point processed is the first in the list, the last point processed is the last in the list. Clusters can be extracted from this list by identifying valleys in the points' reachability, e.g. by using a threshold value. If a maximum distance is specified, this is used to identify clusters, otherwise each separated network will constitute a cluster.
The OPTICS algorithm uses each yet unprocessed point to start a new cluster. The order of the input points is arbitrary and can thus influence the resultant clusters.
optics2 method
EXPERIMENTAL This method is similar to OPTICS, minimizing the reachability of each point. Points are reconnected if their reachability can be reduced. Contrary to OPTICS, a cluster's seed is not fixed but changed if possible. Each point is connected to another point until the core of the cluster (seed point) is reached. Effectively, the initial seed is updated in the process. Thus separated networks of points are created, with each network representing a cluster. The maximum distance is not used.
NOTES
By default, cluster IDs are stored as category values of the points in layer 2.
EXAMPLE
Analysis of random points for areas in areas of the vector urbanarea (North Carolina sample dataset).
First generate 1000 random points within the areas the vector urbanarea and within the subregion, then do clustering and visualize the result:
# pick a subregion of the vector urbanarea
g.region -p n=272950 s=188330 w=574720 e=703090 res=10
# create random points in areas
v.random output=random_points npoints=1000 restrict=urbanarea
# identify clusters
v.cluster input=random_points output=clusters_optics method=optics
# set random vector color table for the clusters
v.colors map=clusters_optics layer=2 use=cat color=random
# display in command line
d.mon wx0
# note the second layer and transparent (none) color of the circle border
d.vect map=clusters_optics layer=2 icon=basic/point size=10 color=none
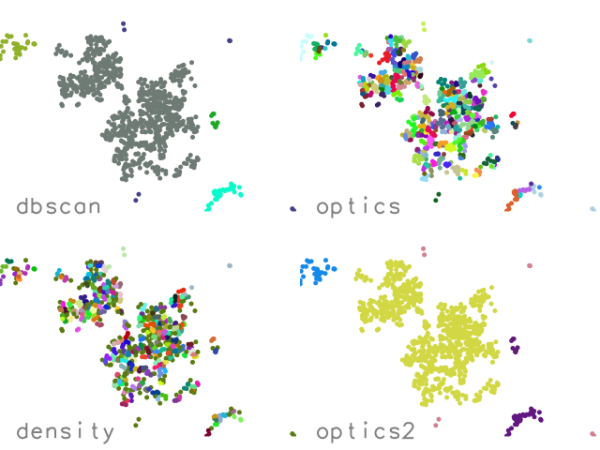
Figure: Four different methods with default settings applied to 1000
random points generated in the same way as in the example.
Generate random points for analysis (100 points per area), use different method for clustering and visualize using color stored the attribute table.
# pick a subregion of the vector urbanarea
g.region -p n=272950 s=188330 w=574720 e=703090 res=10
# create clustered points
v.random output=rand_clust npoints=100 restrict=urbanarea -a
# identify clusters
v.cluster in=rand_clust out=rand_clusters method=dbscan
# create colors for clusters
v.db.addtable map=rand_clusters layer=2 columns="cat integer,grassrgb varchar(11)"
v.colors map=rand_clusters layer=2 use=cat color=random rgb_column=grassrgb
# display with your preferred method
# remember to use the second layer and RGB column
# for example use
d.vect map=rand_clusters layer=2 color=none rgb_column=grassrgb icon=basic/circle
SEE ALSO
AUTHOR
Markus Metz
SOURCE CODE
Available at: v.cluster source code
(history)
Latest change: Wednesday May 07 13:01:38 2025 in commit f20dbc6